The diagnostic accuracy of the GenoType(®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs
- PMID: 25353401
- PMCID: PMC4448219
- DOI: 10.1002/14651858.CD010705.pub2
The diagnostic accuracy of the GenoType(®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs
Abstract
Background: Accurate and rapid tests for tuberculosis (TB) drug resistance are critical for improving patient care and decreasing the transmission of drug-resistant TB. Genotype(®)MTBDRsl (MTBDRsl) is the only commercially-available molecular test for detecting resistance in TB to the fluoroquinolones (FQs; ofloxacin, moxifloxacin and levofloxacin) and the second-line injectable drugs (SLIDs; amikacin, kanamycin and capreomycin), which are used to treat patients with multidrug-resistant (MDR-)TB.
Objectives: To obtain summary estimates of the diagnostic accuracy of MTBDRsl for FQ resistance, SLID resistance and extensively drug-resistant TB (XDR-TB; defined as MDR-TB plus resistance to a FQ and a SLID) when performed (1) indirectly (ie on culture isolates confirmed as TB positive) and (2) directly (ie on smear-positive sputum specimens).To compare summary estimates of the diagnostic accuracy of MTBDRsl for FQ resistance, SLID resistance and XDR-TB by type of testing (indirect versus direct testing).The populations of interest were adults with drug-susceptible TB or drug-resistant TB. The settings of interest were intermediate and central laboratories.
Search methods: We searched the following databases without any language restriction up to 30 January 2014: Cochrane Infectious Diseases Group Specialized Register; MEDLINE; EMBASE; ISI Web of Knowledge; MEDION; LILACS; BIOSIS; SCOPUS; the metaRegister of Controlled Trials; the search portal of the World Health Organization International Clinical Trials Registry Platform; and ProQuest Dissertations & Theses A&I.
Selection criteria: We included all studies that determined MTBDRsl accuracy against a defined reference standard (culture-based drug susceptibility testing (DST), genetic testing or both). We included cross-sectional and diagnostic case-control studies. We excluded unpublished data and conference proceedings.
Data collection and analysis: For each study, two review authors independently extracted data using a standardized form and assessed study quality using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool. We performed meta-analyses to estimate the pooled sensitivity and specificity of MTBDRsl for FQ resistance, SLID resistance, and XDR-TB. We explored the influence of different reference standards. We performed the majority of analyses using a bivariate random-effects model against culture-based DST as the reference standard.
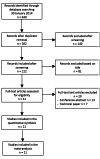
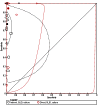
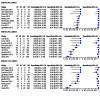
Main results: We included 21 unique studies: 14 studies reported the accuracy of MTBDRsl when done directly, five studies when done indirectly and two studies that did both. Of the 21 studies, 15 studies (71%) were cross-sectional and 11 studies (58%) were located in low-income or middle-income countries. All studies but two were written in English. Nine (43%) of the 21 included studies had a high risk of bias for patient selection. At least half of the studies had low risk of bias for the other QUADAS-2 domains.As a test for FQ resistance measured against culture-based DST, the pooled sensitivity of MTBDRsl when performed indirectly was 83.1% (95% confidence interval (CI) 78.7% to 86.7%) and the pooled specificity was 97.7% (95% CI 94.3% to 99.1%), respectively (16 studies, 1766 participants; 610 confirmed cases of FQ-resistant TB; moderate quality evidence). When performed directly, the pooled sensitivity was 85.1% (95% CI 71.9% to 92.7%) and the pooled specificity was 98.2% (95% CI 96.8% to 99.0%), respectively (seven studies, 1033 participants; 230 confirmed cases of FQ-resistant TB; moderate quality evidence). For indirect testing for FQ resistance, four (0.2%) of 1766 MTBDRsl results were indeterminate, whereas for direct testing 20 (1.9%) of 1033 were MTBDRsl indeterminate (P < 0.001).As a test for SLID resistance measured against culture-based DST, the pooled sensitivity of MTBDRsl when performed indirectly was 76.9% (95% CI 61.1% to 87.6%) and the pooled specificity was 99.5% (95% CI 97.1% to 99.9%), respectively (14 studies, 1637 participants; 414 confirmed cases of SLID-resistant TB; moderate quality evidence). For amikacin resistance, the pooled sensitivity and specificity were 87.9% (95% CI 82.1% to 92.0%) and 99.5% (95% CI 97.5% to 99.9%), respectively. For kanamycin resistance, the pooled sensitivity and specificity were 66.9% (95% CI 44.1% to 83.8%) and 98.6% (95% CI 96.1% to 99.5%), respectively. For capreomycin resistance, the pooled sensitivity and specificity were 79.5% (95% CI 58.3% to 91.4%) and 95.8% (95% CI 93.4% to 97.3%), respectively. When performed directly, the pooled sensitivity for SLID resistance was 94.4% (95% CI 25.2% to 99.9%) and the pooled specificity was 98.2% (95% CI 88.9% to 99.7%), respectively (six studies, 947 participants; 207 confirmed cases of SLID-resistant TB, 740 SLID susceptible cases of TB; very low quality evidence). For indirect testing for SLID resistance, three (0.4%) of 774 MTBDRsl results were indeterminate, whereas for direct testing 53 (6.1%) of 873 were MTBDRsl indeterminate (P < 0.001).As a test for XDR-TB measured against culture-based DST, the pooled sensitivity of MTBDRsl when performed indirectly was 70.9% (95% CI 42.9% to 88.8%) and the pooled specificity was 98.8% (95% CI 96.1% to 99.6%), respectively (eight studies, 880 participants; 173 confirmed cases of XDR-TB; low quality evidence).
Authors' conclusions: In adults with TB, a positive MTBDRsl result for FQ resistance, SLID resistance, or XDR-TB can be treated with confidence. However, MTBDRsl does not detect approximately one in five cases of FQ-resistant TB, and does not detect approximately one in four cases of SLID-resistant TB. Of the three SLIDs, MTBDRsl has the poorest sensitivity for kanamycin resistance. MTBDRsl will miss between one in four and one in three cases of XDR-TB. The diagnostic accuracy of MTBDRsl is similar when done using either culture isolates or smear-positive sputum. As the location of the resistance causing mutations can vary on a strain-by-strain basis, further research is required on test accuracy in different settings and, if genetic sequencing is used as a reference standard, it should examine all resistance-determining regions. Given the confidence one can have in a positive result, and the ability of the test to provide results within a matter of days, MTBDRsl may be used as an initial test for second-line drug resistance. However, when the test reports a negative result, clinicians may still wish to carry out conventional testing.
Figures






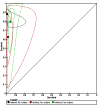

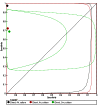


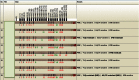


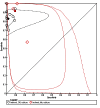


























References
-
- Barnard M, Warren R, Gey Van Pittius N, van Helden P, Bosman M, Streicher E, et al. Genotype MTBDRsl line probe assay shortens time to diagnosis of extensively drug-resistant tuberculosis in a high-throughput diagnostic laboratory. American Journal of Respiratory and Critical Care Medicine. 2012;186(12):1298–305. - PubMed
-
- Chikamatsu K, Aono A, Yamada H, Mitarai S. Evaluation of GenoType MTBDRsl for testing resistance of Mycobacterium tuberculosis isolates to fluoroquinlone, aminoglycoside, and ethambutol. Kekkaku (Tuberculosis) 2012;87(10):641–7. - PubMed
-
- Fan QW, Guo J, Zhang HZ, Wu XY, Hu XN, Qian XQ, et al. The characteristics of drug resistant relevant genes in multidrug-resistant and extensively drug-resistant tuberculosis by fast molecular assay [Chinese] Chinese Journal of Microbiology and Immunology. 2011;31(12):1133–7.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

