Integrative data mining highlights candidate genes for monogenic myopathies
- PMID: 25353622
- PMCID: PMC4213015
- DOI: 10.1371/journal.pone.0110888
Integrative data mining highlights candidate genes for monogenic myopathies
Erratum in
-
Correction: integrative data mining highlights candidate genes for monogenic myopathies.PLoS One. 2015 Feb 3;10(2):e0117884. doi: 10.1371/journal.pone.0117884. eCollection 2015. PLoS One. 2015. PMID: 25646818 Free PMC article.
Abstract
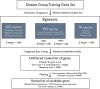
Inherited myopathies are a heterogeneous group of disabling disorders with still barely understood pathological mechanisms. Around 40% of afflicted patients remain without a molecular diagnosis after exclusion of known genes. The advent of high-throughput sequencing has opened avenues to the discovery of new implicated genes, but a working list of prioritized candidate genes is necessary to deal with the complexity of analyzing large-scale sequencing data. Here we used an integrative data mining strategy to analyze the genetic network linked to myopathies, derive specific signatures for inherited myopathy and related disorders, and identify and rank candidate genes for these groups. Training sets of genes were selected after literature review and used in Manteia, a public web-based data mining system, to extract disease group signatures in the form of enriched descriptor terms, which include functional annotation, human and mouse phenotypes, as well as biological pathways and protein interactions. These specific signatures were then used as an input to mine and rank candidate genes, followed by filtration against skeletal muscle expression and association with known diseases. Signatures and identified candidate genes highlight both potential common pathological mechanisms and allelic disease groups. Recent discoveries of gene associations to diseases, like B3GALNT2, GMPPB and B3GNT1 to congenital muscular dystrophies, were prioritized in the ranked lists, suggesting a posteriori validation of our approach and predictions. We show an example of how the ranked lists can be used to help analyze high-throughput sequencing data to identify candidate genes, and highlight the best candidate genes matching genomic regions linked to myopathies without known causative genes. This strategy can be automatized to generate fresh candidate gene lists, which help cope with database annotation updates as new knowledge is incorporated.
Conflict of interest statement
Figures


References
-
- Kaplan JC, Hamroun D (2013) The 2014 version of the gene table of monogenic neuromuscular disorders (nuclear genome). Neuromuscul Disord 23: 1081–1111. - PubMed
-
- Mercuri E, Muntoni F (2013) Muscular dystrophies. Lancet 381: 845–860. - PubMed
-
- Mercuri E, Muntoni F (2012) The ever-expanding spectrum of congenital muscular dystrophies. Ann Neurol 72: 9–17. - PubMed
-
- Heatwole CR, Statland JM, Logigian EL (2013) The diagnosis and treatment of myotonic disorders. Muscle Nerve 47: 632–648. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

