The E3 ubiquitin ligase thyroid hormone receptor-interacting protein 12 targets pancreas transcription factor 1a for proteasomal degradation
- PMID: 25355311
- PMCID: PMC4271242
- DOI: 10.1074/jbc.M114.620104
The E3 ubiquitin ligase thyroid hormone receptor-interacting protein 12 targets pancreas transcription factor 1a for proteasomal degradation
Abstract
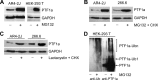
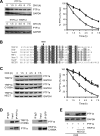
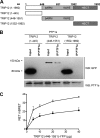
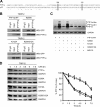
Pancreas transcription factor 1a (PTF1a) plays a crucial role in the early development of the pancreas and in the maintenance of the acinar cell phenotype. Several transcriptional mechanisms regulating expression of PTF1a have been identified. However, regulation of PTF1a protein stability and degradation is still unexplored. Here, we report that inhibition of proteasome leads to elevated levels of PTF1a and to the existence of polyubiquitinated forms of PTF1a. We used the Sos recruitment system, an alternative two-hybrid system method to detect protein-protein interactions in the cytoplasm and to map the interactome of PTF1a. We identified TRIP12 (thyroid hormone receptor-interacting protein 12), an E3 ubiquitin-protein ligase as a new partner of PTF1a. We confirmed PTF1a/TRIP12 interaction in acinar cell lines and in co-transfected HEK-293T cells. The protein stability of PTF1a is significantly increased upon decreased expression of TRIP12. It is reduced upon overexpression of TRIP12 but not a catalytically inactive TRIP12-C1959A mutant. We identified a region of TRIP12 required for interaction and identified lysine 312 of PTF1a as essential for proteasomal degradation. We also demonstrate that TRIP12 down-regulates PTF1a transcriptional and antiproliferative activities. Our data suggest that an increase in TRIP12 expression can play a part in PTF1a down-regulation and indicate that PTF1a/TRIP12 functional interaction may regulate pancreatic epithelial cell homeostasis.
Keywords: Basic Helix-Loop-Helix Transcription Factor (bHLH); E3 Ubiquitin Ligase; Pancreas; Pancreatic Cancer; Proteasome; Protein Degradation; Protein-Protein Interaction; Transcription Factor; Ubiquitylation (Ubiquitination); Yeast Two-Hybrid.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
The E3 ubiquitin ligase TRIP12 is required for pancreatic acinar cell plasticity and pancreatic carcinogenesis.J Pathol. 2024 Aug;263(4-5):466-481. doi: 10.1002/path.6298. Epub 2024 Jun 25. J Pathol. 2024. PMID: 38924548
-
Proteasomal degradation of the tumour suppressor FBW7 requires branched ubiquitylation by TRIP12.Nat Commun. 2021 Apr 6;12(1):2043. doi: 10.1038/s41467-021-22319-5. Nat Commun. 2021. PMID: 33824312 Free PMC article.
-
Trip12 is an E3 ubiquitin ligase for USP7/HAUSP involved in the DNA damage response.FEBS Lett. 2016 Dec;590(23):4213-4222. doi: 10.1002/1873-3468.12471. Epub 2016 Nov 11. FEBS Lett. 2016. PMID: 27800609
-
E3 Ubiquitin Ligase TRIP12: Regulation, Structure, and Physiopathological Functions.Int J Mol Sci. 2020 Nov 12;21(22):8515. doi: 10.3390/ijms21228515. Int J Mol Sci. 2020. PMID: 33198194 Free PMC article. Review.
-
Design Principles Involving Protein Disorder Facilitate Specific Substrate Selection and Degradation by the Ubiquitin-Proteasome System.J Biol Chem. 2016 Mar 25;291(13):6723-31. doi: 10.1074/jbc.R115.692665. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851277 Free PMC article. Review.
Cited by
-
Transcription factor Ptf1a in development, diseases and reprogramming.Cell Mol Life Sci. 2019 Mar;76(5):921-940. doi: 10.1007/s00018-018-2972-z. Epub 2018 Nov 23. Cell Mol Life Sci. 2019. PMID: 30470852 Free PMC article. Review.
-
The oncogenic E3 ligase TRIP12 suppresses epithelial-mesenchymal transition (EMT) and mesenchymal traits through ZEB1/2.Cell Death Discov. 2021 May 7;7(1):95. doi: 10.1038/s41420-021-00479-z. Cell Death Discov. 2021. PMID: 33963176 Free PMC article.
-
Cytoplasmic Localization of Thyroid Hormone Receptor (TR) Alpha and Nuclear Expression of Its Isoform TRα2 Determine Survival in Breast Cancer in Opposite Ways.Cancers (Basel). 2023 Jul 13;15(14):3610. doi: 10.3390/cancers15143610. Cancers (Basel). 2023. PMID: 37509273 Free PMC article.
-
The E3 ubiquitin ligase TRIP12 participates in cell cycle progression and chromosome stability.Sci Rep. 2020 Jan 21;10(1):789. doi: 10.1038/s41598-020-57762-9. Sci Rep. 2020. PMID: 31964993 Free PMC article.
-
Five enzymes of the Arg/N-degron pathway form a targeting complex: The concept of superchanneling.Proc Natl Acad Sci U S A. 2020 May 19;117(20):10778-10788. doi: 10.1073/pnas.2003043117. Epub 2020 May 4. Proc Natl Acad Sci U S A. 2020. PMID: 32366662 Free PMC article.
References
-
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 - PubMed
-
- Lee J. W., Choi H. S., Gyuris J., Brent R., Moore D. D. (1995) Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 9, 243–254 - PubMed
-
- Kajiro M., Tsuchiya M., Kawabe Y., Furumai R., Iwasaki N., Hayashi Y., Katano M., Nakajima Y., Goto N., Watanabe T., Murayama A., Oishi H., Ema M., Takahashi S., Kishimoto H., Yanagisawa J. (2011) The E3 ubiquitin ligase activity of Trip12 is essential for mouse embryogenesis. PLoS One 6, e25871. - PMC - PubMed
-
- Park Y., Yoon S. K., Yoon J. B. (2009) The HECT domain of TRIP12 ubiquitinates substrates of the ubiquitin fusion degradation pathway. J. Biol. Chem. 284, 1540–1549 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

