Derivatives of mesoxalic acid block translocation of HIV-1 reverse transcriptase
- PMID: 25355312
- PMCID: PMC4340395
- DOI: 10.1074/jbc.M114.614305
Derivatives of mesoxalic acid block translocation of HIV-1 reverse transcriptase
Abstract
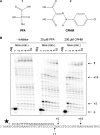
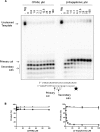
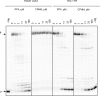
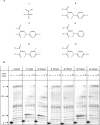
The pyrophosphate mimic and broad spectrum antiviral phosphonoformic acid (PFA, foscarnet) was shown to freeze the pre-translocational state of the reverse transcriptase (RT) complex of the human immunodeficiency virus type 1 (HIV-1). However, PFA lacks a specificity domain, which is seen as a major reason for toxic side effects associated with the clinical use of this drug. Here, we studied the mechanism of inhibition of HIV-1 RT by the 4-chlorophenylhydrazone of mesoxalic acid (CPHM) and demonstrate that this compound also blocks RT translocation. Hot spots for inhibition with PFA or CPHM occur at template positions with a bias toward pre-translocation. Mutations at active site residue Asp-185 compromise binding of both compounds. Moreover, divalent metal ions are required for the formation of ternary complexes with either of the two compounds. However, CPHM contains both an anchor domain that likely interacts with the catalytic metal ions and a specificity domain. Thus, although the inhibitor binding sites may partly overlap, they are not identical. The K65R mutation in HIV-1 RT, which reduces affinity to PFA, increases affinity to CPHM. Details with respect to the binding sites of the two inhibitors are provided on the basis of mutagenesis studies, structure-activity relationship analyses with newly designed CPHM derivatives, and in silico docking experiments. Together, these findings validate the pre-translocated complex of HIV-1 RT as a specific target for the development of novel classes of RT inhibitors.
Keywords: Antiviral Agent; Drug Action; Drug Resistance; Human Immunodeficiency Virus (HIV); Reverse Transcription.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
Inhibition of HIV-1 reverse transcriptase-catalyzed DNA strand transfer reactions by 4-chlorophenylhydrazone of mesoxalic acid.Biochemistry. 2000 Nov 21;39(46):14279-91. doi: 10.1021/bi0015764. Biochemistry. 2000. PMID: 11087377
-
The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation.J Biol Chem. 2007 Feb 2;282(5):3337-46. doi: 10.1074/jbc.M607710200. Epub 2006 Nov 30. J Biol Chem. 2007. PMID: 17145704
-
Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain.Chem Biol Drug Des. 2012 Nov;80(5):706-16. doi: 10.1111/cbdd.12010. Epub 2012 Aug 31. Chem Biol Drug Des. 2012. PMID: 22846652 Free PMC article.
-
Cutting into the Substrate Dominance: Pharmacophore and Structure-Based Approaches toward Inhibiting Human Immunodeficiency Virus Reverse Transcriptase-Associated Ribonuclease H.Acc Chem Res. 2020 Jan 21;53(1):218-230. doi: 10.1021/acs.accounts.9b00450. Epub 2019 Dec 27. Acc Chem Res. 2020. PMID: 31880912 Free PMC article. Review.
-
Evolving understanding of HIV-1 reverse transcriptase structure, function, inhibition, and resistance.Curr Opin Struct Biol. 2020 Apr;61:113-123. doi: 10.1016/j.sbi.2019.11.011. Epub 2020 Jan 11. Curr Opin Struct Biol. 2020. PMID: 31935541 Free PMC article. Review.
Cited by
-
Pronounced Inhibition Shift from HIV Reverse Transcriptase to Herpetic DNA Polymerases by Increasing the Flexibility of α-Carboxy Nucleoside Phosphonates.J Med Chem. 2015 Oct 22;58(20):8110-27. doi: 10.1021/acs.jmedchem.5b01180. Epub 2015 Oct 9. J Med Chem. 2015. PMID: 26450273 Free PMC article.
-
Long-acting drugs and formulations for the treatment and prevention of HIV infection.Int J Antimicrob Agents. 2021 Jan;57(1):106220. doi: 10.1016/j.ijantimicag.2020.106220. Epub 2020 Nov 6. Int J Antimicrob Agents. 2021. PMID: 33166693 Free PMC article. Review.
-
Long-acting implants to treat and prevent HIV infection.Curr Opin HIV AIDS. 2020 Jan;15(1):33-41. doi: 10.1097/COH.0000000000000591. Curr Opin HIV AIDS. 2020. PMID: 31764198 Free PMC article.
-
Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.ACS Chem Biol. 2016 Aug 19;11(8):2158-64. doi: 10.1021/acschembio.6b00187. Epub 2016 Jun 6. ACS Chem Biol. 2016. PMID: 27192549 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

