Yeast 14-3-3 protein functions as a comodulator of transcription by inhibiting coactivator functions
- PMID: 25355315
- PMCID: PMC4271238
- DOI: 10.1074/jbc.M114.592287
Yeast 14-3-3 protein functions as a comodulator of transcription by inhibiting coactivator functions
Abstract
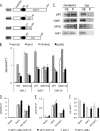
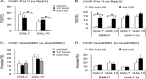
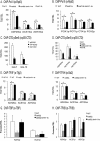
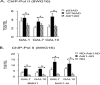
In eukaryotes combinatorial activation of transcription is an important component of gene regulation. In the budding yeast Saccharomyces cerevisiae, Adr1-Cat8 and Adr1-Oaf1/Pip2 are pairs of activators that act together to regulate two diverse sets of genes. Transcription activation of both sets is regulated positively by the yeast AMP-activated protein kinase homolog, Snf1, in response to low glucose or the presence of a non-fermentable carbon source and negatively by two redundant 14-3-3 isoforms, Bmh1 and Bmh2. Bmh regulates the function of these pairs at a post-promoter binding step by direct binding to Adr1. However, how Bmh regulates transcription after activator binding remains unknown. In the present study we analyzed Bmh-mediated regulation of two sets of genes activated independently by these pairs of activators. We report that Bmh inhibits mRNA synthesis when the second activator is absent. Using gene fusions we show that Bmh binding to the Adr1 regulatory domain inhibits an Adr1 activation domain but not a heterologous activation domain or artificially recruited Mediator, consistent with Bmh acting at a step in transcription downstream of activator binding. Bmh inhibits the assembly and the function of a preinitiation complex (PIC). Gene expression studies suggest that Bmh regulates Adr1 activity through the coactivators Mediator and Swi/Snf. Mediator recruitment appeared to occur normally, but PIC formation and function were defective, suggesting that Bmh inhibits a step between Mediator recruitment and PIC activation.
Keywords: 14-3-3 Protein; Adr1; Cat8; Combinatorial Regulation; Gene Regulation; Gene Transcription; Oaf1/Pip2; RNA Polymerase II; Transcription Coactivator.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae.Mol Cell Biol. 2013 Feb;33(4):712-24. doi: 10.1128/MCB.01226-12. Epub 2012 Dec 3. Mol Cell Biol. 2013. PMID: 23207903 Free PMC article.
-
Snf1 dependence of peroxisomal gene expression is mediated by Adr1.J Biol Chem. 2010 Apr 2;285(14):10703-14. doi: 10.1074/jbc.M109.079848. Epub 2010 Feb 6. J Biol Chem. 2010. PMID: 20139423 Free PMC article.
-
14-3-3 (Bmh) proteins inhibit transcription activation by Adr1 through direct binding to its regulatory domain.Mol Cell Biol. 2010 Nov;30(22):5273-83. doi: 10.1128/MCB.00715-10. Epub 2010 Sep 20. Mol Cell Biol. 2010. PMID: 20855531 Free PMC article.
-
Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae.Curr Genet. 2003 Jun;43(3):139-60. doi: 10.1007/s00294-003-0381-8. Epub 2003 Apr 25. Curr Genet. 2003. PMID: 12715202 Review.
-
Transcriptional regulation of nonfermentable carbon utilization in budding yeast.FEMS Yeast Res. 2010 Feb;10(1):2-13. doi: 10.1111/j.1567-1364.2009.00555.x. Epub 2009 Jul 18. FEMS Yeast Res. 2010. PMID: 19686338 Free PMC article. Review.
Cited by
-
SWI/SNF senses carbon starvation with a pH-sensitive low-complexity sequence.Elife. 2022 Feb 7;11:e70344. doi: 10.7554/eLife.70344. Elife. 2022. PMID: 35129437 Free PMC article.
-
A ubiquitination-mediated degradation system to target 14-3-3-binding phosphoproteins.Heliyon. 2023 May 20;9(5):e16318. doi: 10.1016/j.heliyon.2023.e16318. eCollection 2023 May. Heliyon. 2023. PMID: 37251884 Free PMC article.
-
Regulation of Rim4 distribution, function, and stability during meiosis by PKA, Cdc14, and 14-3-3 proteins.Cell Rep. 2023 Sep 26;42(9):113052. doi: 10.1016/j.celrep.2023.113052. Epub 2023 Sep 1. Cell Rep. 2023. PMID: 37659077 Free PMC article.
-
Functional analysis of Paracoccidioides brasiliensis 14-3-3 adhesin expressed in Saccharomyces cerevisiae.BMC Microbiol. 2015 Nov 4;15:256. doi: 10.1186/s12866-015-0586-2. BMC Microbiol. 2015. PMID: 26537993 Free PMC article.
-
Snf1-Dependent Transcription Confers Glucose-Induced Decay upon the mRNA Product.Mol Cell Biol. 2015 Dec 14;36(4):628-44. doi: 10.1128/MCB.00436-15. Print 2016 Feb 15. Mol Cell Biol. 2015. PMID: 26667037 Free PMC article.
References
-
- Glass C. K. (1994) Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr. Rev. 15, 391–407 - PubMed
-
- Shore P., Sharrocks A. D. (1995) The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13 - PubMed
-
- Herr W., Cleary M. A. (1995) The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 9, 1679–1693 - PubMed
-
- Messenguy F., Dubois E. (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316, 1–21 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

