DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain
- PMID: 25355443
- PMCID: PMC4610814
- DOI: 10.1002/ajmg.b.32278
DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain
Abstract
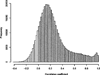
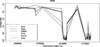
DNA methylation has become increasingly recognized in the etiology of psychiatric disorders. Because brain tissue is not accessible in living humans, epigenetic studies are most often conducted in blood. Saliva is often collected for genotyping studies but is rarely used to examine DNA methylation because the proportion of epithelial cells and leukocytes varies extensively between individuals. The goal of this study was to evaluate whether saliva DNA is informative for studies of psychiatric disorders. DNA methylation (HumanMethylation450 BeadChip) was assessed in saliva and blood samples from 64 adult African Americans. Analyses were conducted using linear regression adjusted for appropriate covariates, including estimated cellular proportions. DNA methylation from brain tissues (cerebellum, frontal cortex, entorhinal cortex, and superior temporal gyrus) was obtained from a publically available dataset. Saliva and blood methylation was clearly distinguishable though there was positive correlation overall. There was little correlation in CpG sites within relevant candidate genes. Correlated CpG sites were more likely to occur in areas of low CpG density (i.e., CpG shores and open seas). There was more variability in CpG sites from saliva than blood, which may reflect its heterogeneity. Finally, DNA methylation in saliva appeared more similar to patterns from each of the brain regions examined overall than methylation in blood. Thus, this study provides a framework for using DNA methylation from saliva and suggests that DNA methylation of saliva may offer distinct opportunities for epidemiological and longitudinal studies of psychiatric traits.
Keywords: EWAS; HumanMethylation450; biomarker; epigenetic; oragene.
© 2014 Wiley Periodicals, Inc.
Figures


References
-
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, Harada A, Hultman CM, Sullivan PF, Magnusson PK, van den Oord EJ. Methylome-wide association study of schizophrenia: Identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71(3):255–264. - PMC - PubMed
-
- Beyan H, Down TA, Ramagopalan SV, Uvebrant K, Nilsson A, Holland ML, Gemma C, Giovannoni G, Boehm BO, Ebers GC, Lernmark A, Cilio CM, Leslie RD, Rakyan VK. Guthrie card methylomics identifies temporally stable epialleles that ate present at birth in humans. Genome Res. 2012;22(11):2138–2145. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 MH071537/MH/NIMH NIH HHS/United States
- K01 MH085806/MH/NIMH NIH HHS/United States
- R01 MH071537/MH/NIMH NIH HHS/United States
- R01 MH096764/MH/NIMH NIH HHS/United States
- MH071537/MH/NIMH NIH HHS/United States
- MH085806/MH/NIMH NIH HHS/United States
- R01 HD071982/HD/NICHD NIH HHS/United States
- HD071982/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 MH094757/MH/NIMH NIH HHS/United States
- 281338/ERC_/European Research Council/International
- MH096764/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

