Endonuclease G preferentially cleaves 5-hydroxymethylcytosine-modified DNA creating a substrate for recombination
- PMID: 25355512
- PMCID: PMC4245937
- DOI: 10.1093/nar/gku1032
Endonuclease G preferentially cleaves 5-hydroxymethylcytosine-modified DNA creating a substrate for recombination
Abstract
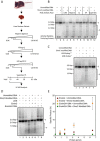
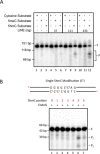
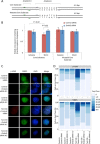
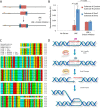
5-hydroxymethylcytosine (5hmC) has been suggested to be involved in various nucleic acid transactions and cellular processes, including transcriptional regulation, demethylation of 5-methylcytosine and stem cell pluripotency. We have identified an activity that preferentially catalyzes the cleavage of double-stranded 5hmC-modified DNA. Using biochemical methods we purified this activity from mouse liver extracts and demonstrate that the enzyme responsible for the cleavage of 5hmC-modified DNA is Endonuclease G (EndoG). We show that recombinant EndoG preferentially recognizes and cleaves a core sequence when one specific cytosine within that core sequence is hydroxymethylated. Additionally, we provide in vivo evidence that EndoG catalyzes the formation of double-stranded DNA breaks and that this cleavage is dependent upon the core sequence, EndoG and 5hmC. Finally, we demonstrate that the 5hmC modification can promote conservative recombination in an EndoG-dependent manner.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.Nucleic Acids Res. 2020 Apr 17;48(7):3962-3974. doi: 10.1093/nar/gkaa117. Nucleic Acids Res. 2020. PMID: 32095813 Free PMC article.
-
Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.Nucleic Acids Res. 2014 Jul;42(12):7947-59. doi: 10.1093/nar/gku497. Epub 2014 Jun 3. Nucleic Acids Res. 2014. PMID: 24895434 Free PMC article.
-
Endonuclease G plays a role in immunoglobulin class switch DNA recombination by introducing double-strand breaks in switch regions.Mol Immunol. 2011 Jan;48(4):610-22. doi: 10.1016/j.molimm.2010.10.023. Epub 2010 Nov 26. Mol Immunol. 2011. PMID: 21111482 Free PMC article.
-
Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma.Cell Prolif. 2019 Jul;52(4):e12626. doi: 10.1111/cpr.12626. Epub 2019 Apr 29. Cell Prolif. 2019. PMID: 31033072 Free PMC article. Review.
-
5-Hydroxymethylcytosine: a stable or transient DNA modification?Genomics. 2014 Nov;104(5):314-23. doi: 10.1016/j.ygeno.2014.08.015. Epub 2014 Aug 30. Genomics. 2014. PMID: 25181633 Free PMC article. Review.
Cited by
-
Cardiomyocyte hypertrophy induced by Endonuclease G deficiency requires reactive oxygen radicals accumulation and is inhibitable by the micropeptide humanin.Redox Biol. 2018 Jun;16:146-156. doi: 10.1016/j.redox.2018.02.021. Epub 2018 Mar 1. Redox Biol. 2018. PMID: 29502044 Free PMC article.
-
Endonuclease G promotes mitochondrial genome cleavage and replication.Oncotarget. 2018 Apr 6;9(26):18309-18326. doi: 10.18632/oncotarget.24822. eCollection 2018 Apr 6. Oncotarget. 2018. PMID: 29719607 Free PMC article.
-
Effect of Hydroxymethylcytosine on the Structure and Stability of Holliday Junctions.Biochemistry. 2016 Oct 18;55(41):5781-5789. doi: 10.1021/acs.biochem.6b00801. Epub 2016 Oct 5. Biochemistry. 2016. PMID: 27653243 Free PMC article.
-
5-hydroxymethylcytosine Marks Mammalian Origins Acting as a Barrier to Replication.Sci Rep. 2019 Jul 30;9(1):11065. doi: 10.1038/s41598-019-47528-3. Sci Rep. 2019. PMID: 31363131 Free PMC article.
-
Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.Nucleic Acids Res. 2020 Apr 17;48(7):3962-3974. doi: 10.1093/nar/gkaa117. Nucleic Acids Res. 2020. PMID: 32095813 Free PMC article.
References
-
- Cannon-Carlson S.V., Gokhale H., Teebor G.W. Purification and characterization of 5-hydroxymethyluracil-DNA glycosylase from calf thymus. Its possible role in the maintenance of methylated cytosine residues. J. Biol. Chem. 1989;264:13306–13312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

