Efficient processing of abasic sites by bacterial nonhomologous end-joining Ku proteins
- PMID: 25355514
- PMCID: PMC4245934
- DOI: 10.1093/nar/gku1029
Efficient processing of abasic sites by bacterial nonhomologous end-joining Ku proteins
Abstract
Intracellular reactive oxygen species as well as the exposure to harsh environmental conditions can cause, in the single chromosome of Bacillus subtilis spores, the formation of apurinic/apyrimidinic (AP) sites and strand breaks whose repair during outgrowth is crucial to guarantee cell viability. Whereas double-stranded breaks are mended by the nonhomologous end joining (NHEJ) system composed of an ATP-dependent DNA Ligase D (LigD) and the DNA-end-binding protein Ku, repair of AP sites would rely on an AP endonuclease or an AP-lyase, a polymerase and a ligase. Here we show that B. subtilis Ku (BsuKu), along with its pivotal role in allowing joining of two broken ends by B. subtilis LigD (BsuLigD), is endowed with an AP/deoxyribose 5'-phosphate (5'-dRP)-lyase activity that can act on ssDNA, nicked molecules and DNA molecules without ends, suggesting a potential role in BER during spore outgrowth. Coordination with BsuLigD makes possible the efficient joining of DNA ends with near terminal abasic sites. The role of this new enzymatic activity of Ku and its potential importance in the NHEJ pathway is discussed. The presence of an AP-lyase activity also in the homolog protein from the distantly related bacterium Pseudomonas aeruginosa allows us to expand our results to other bacterial Ku proteins.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

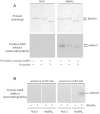





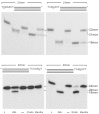

Similar articles
-
Identification of a conserved 5'-dRP lyase activity in bacterial DNA repair ligase D and its potential role in base excision repair.Nucleic Acids Res. 2016 Feb 29;44(4):1833-44. doi: 10.1093/nar/gkw054. Epub 2016 Jan 29. Nucleic Acids Res. 2016. PMID: 26826709 Free PMC article.
-
Bacterial Ligase D preternary-precatalytic complex performs efficient abasic sites processing at double strand breaks during nonhomologous end joining.Nucleic Acids Res. 2019 Jun 4;47(10):5276-5292. doi: 10.1093/nar/gkz265. Nucleic Acids Res. 2019. PMID: 30976810 Free PMC article.
-
Ku is a 5'-dRP/AP lyase that excises nucleotide damage near broken ends.Nature. 2010 Apr 22;464(7292):1214-7. doi: 10.1038/nature08926. Epub 2010 Apr 11. Nature. 2010. PMID: 20383123 Free PMC article.
-
[The role of Ku antigen in the repair of apurinic/apyrimidinic sites in DNA].Mol Biol (Mosk). 2015 Jan-Feb;49(1):67-74. Mol Biol (Mosk). 2015. PMID: 25916111 Review. Russian.
-
End-processing nucleases and phosphodiesterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair.DNA Repair (Amst). 2016 Jul;43:57-68. doi: 10.1016/j.dnarep.2016.05.011. Epub 2016 May 18. DNA Repair (Amst). 2016. PMID: 27262532 Review.
Cited by
-
Identification of a conserved 5'-dRP lyase activity in bacterial DNA repair ligase D and its potential role in base excision repair.Nucleic Acids Res. 2016 Feb 29;44(4):1833-44. doi: 10.1093/nar/gkw054. Epub 2016 Jan 29. Nucleic Acids Res. 2016. PMID: 26826709 Free PMC article.
-
The Mycobacterium tuberculosis Ku C-terminus is a multi-purpose arm for binding DNA and LigD and stimulating ligation.Nucleic Acids Res. 2022 Oct 28;50(19):11040-11057. doi: 10.1093/nar/gkac906. Nucleic Acids Res. 2022. PMID: 36250639 Free PMC article.
-
Multiple and Variable NHEJ-Like Genes Are Involved in Resistance to DNA Damage in Streptomyces ambofaciens.Front Microbiol. 2016 Nov 28;7:1901. doi: 10.3389/fmicb.2016.01901. eCollection 2016. Front Microbiol. 2016. PMID: 27965636 Free PMC article.
-
C-terminal region of bacterial Ku controls DNA bridging, DNA threading and recruitment of DNA ligase D for double strand breaks repair.Nucleic Acids Res. 2016 Jun 2;44(10):4785-4806. doi: 10.1093/nar/gkw149. Epub 2016 Mar 9. Nucleic Acids Res. 2016. PMID: 26961308 Free PMC article.
-
The Multifaceted Roles of Ku70/80.Int J Mol Sci. 2021 Apr 16;22(8):4134. doi: 10.3390/ijms22084134. Int J Mol Sci. 2021. PMID: 33923616 Free PMC article. Review.
References
-
- Aguilera A., Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. - PubMed
-
- Daley J.M., Palmbos P.L., Wu D., Wilson T.E. Nonhomologous end joining in yeast. Annu. Rev. Genet. 2005;39:431–451. - PubMed
-
- Pitcher R.S., Brissett N.C., Doherty A.J. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 2007;61:259–282. - PubMed
-
- Shuman S., Glickman M.S. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007;5:852–861. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

