Microevolution of nematode miRNAs reveals diverse modes of selection
- PMID: 25355809
- PMCID: PMC4255771
- DOI: 10.1093/gbe/evu239
Microevolution of nematode miRNAs reveals diverse modes of selection
Abstract
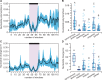
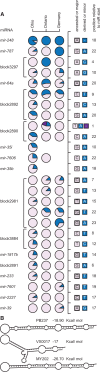
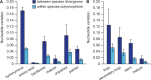
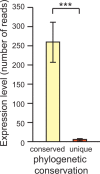
Micro-RNA (miRNA) genes encode abundant small regulatory RNAs that play key roles during development and in homeostasis by fine tuning and buffering gene expression. This layer of regulatory control over transcriptional networks is preserved by selection across deep evolutionary time, yet selection pressures on individual miRNA genes in contemporary populations remain poorly characterized in any organism. Here, we quantify nucleotide variability for 129 miRNAs in the genome of the nematode Caenorhabditis remanei to understand the microevolution of this important class of regulatory genes. Our analysis of three population samples and C. remanei's sister species revealed ongoing natural selection that constrains evolution of all sequence domains within miRNA hairpins. We also show that new miRNAs evolve faster than older miRNAs but that selection nevertheless favors their persistence. Despite the ongoing importance of purging of new mutations, we discover a trove of >400 natural miRNA sequence variants that include single nucleotide polymorphisms in seed motifs, indels that ablate miRNA functional domains, and origination of new miRNAs by duplication. Moreover, we demonstrate substantial nucleotide divergence of pre-miRNA hairpin alleles between populations and sister species. These findings from the first global survey of miRNA microevolution in Caenorhabditis support the idea that changes in gene expression, mediated through divergence in miRNA regulation, can contribute to phenotypic novelty and adaptation to specific environments in the present day as well as the distant past.
Keywords: Caenorhabditis; gene expression; miRNA; nucleotide variation; regulatory networks.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Repertoire and evolution of miRNA genes in four divergent nematode species.Genome Res. 2009 Nov;19(11):2064-74. doi: 10.1101/gr.093781.109. Epub 2009 Sep 15. Genome Res. 2009. PMID: 19755563 Free PMC article.
-
Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification.RNA. 2004 Sep;10(9):1309-22. doi: 10.1261/rna.5206304. RNA. 2004. PMID: 15317971 Free PMC article.
-
MicroRNA sequence variation potentially contributes to within-species functional divergence in the nematode Caenorhabditis briggsae.Genetics. 2011 Nov;189(3):967-76. doi: 10.1534/genetics.111.132795. Epub 2011 Sep 2. Genetics. 2011. PMID: 21890738 Free PMC article.
-
Plant microRNAs: an insight into their gene structures and evolution.Semin Cell Dev Biol. 2010 Oct;21(8):782-9. doi: 10.1016/j.semcdb.2010.07.009. Epub 2010 Aug 4. Semin Cell Dev Biol. 2010. PMID: 20691276 Review.
-
The evolution of microRNAs in plants.Curr Opin Plant Biol. 2017 Feb;35:61-67. doi: 10.1016/j.pbi.2016.11.006. Epub 2016 Nov 22. Curr Opin Plant Biol. 2017. PMID: 27886593 Free PMC article. Review.
Cited by
-
The landscape of extreme genomic variation in the highly adaptable Atlantic killifish.Genome Biol Evol. 2017 Mar;9(3):659-676. doi: 10.1093/gbe/evx023. Epub 2017 Feb 13. Genome Biol Evol. 2017. PMID: 28201664 Free PMC article.
-
Origin, Evolution, and Loss of Bacterial Small RNAs.Microbiol Spectr. 2018 Apr;6(2):10.1128/microbiolspec.rwr-0004-2017. doi: 10.1128/microbiolspec.RWR-0004-2017. Microbiol Spectr. 2018. PMID: 29623872 Free PMC article. Review.
-
Clustering pattern and evolution characteristic of microRNAs in grass carp (Ctenopharyngodon idella).BMC Genomics. 2023 Feb 13;24(1):73. doi: 10.1186/s12864-023-09159-x. BMC Genomics. 2023. PMID: 36782132 Free PMC article.
-
Emergence of New sRNAs in Enteric Bacteria is Associated with Low Expression and Rapid Evolution.J Mol Evol. 2017 Apr;84(4):204-213. doi: 10.1007/s00239-017-9793-9. Epub 2017 Apr 12. J Mol Evol. 2017. PMID: 28405712
-
Dynamic Birth and Death of Argonaute Gene Family Functional Repertoire Across Caenorhabditis Nematodes.Genome Biol Evol. 2025 May 30;17(6):evaf106. doi: 10.1093/gbe/evaf106. Genome Biol Evol. 2025. PMID: 40497303 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

