Multiscale approach to the determination of the photoactive yellow protein signaling state ensemble
- PMID: 25356903
- PMCID: PMC4214557
- DOI: 10.1371/journal.pcbi.1003797
Multiscale approach to the determination of the photoactive yellow protein signaling state ensemble
Erratum in
-
Correction: Multiscale Approach to the Determination of the Photoactive Yellow Protein Signaling State Ensemble.PLoS Comput Biol. 2017 Sep 25;13(9):e1005770. doi: 10.1371/journal.pcbi.1005770. eCollection 2017 Sep. PLoS Comput Biol. 2017. PMID: 28945749 Free PMC article.
Abstract
The nature of the optical cycle of photoactive yellow protein (PYP) makes its elucidation challenging for both experiment and theory. The long transition times render conventional simulation methods ineffective, and yet the short signaling-state lifetime makes experimental data difficult to obtain and interpret. Here, through an innovative combination of computational methods, a prediction and analysis of the biological signaling state of PYP is presented. Coarse-grained modeling and locally scaled diffusion map are first used to obtain a rough bird's-eye view of the free energy landscape of photo-activated PYP. Then all-atom reconstruction, followed by an enhanced sampling scheme; diffusion map-directed-molecular dynamics are used to focus in on the signaling-state region of configuration space and obtain an ensemble of signaling state structures. To the best of our knowledge, this is the first time an all-atom reconstruction from a coarse grained model has been performed in a relatively unexplored region of molecular configuration space. We compare our signaling state prediction with previous computational and more recent experimental results, and the comparison is favorable, which validates the method presented. This approach provides additional insight to understand the PYP photo cycle, and can be applied to other systems for which more direct methods are impractical.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

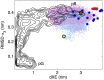
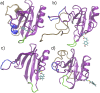
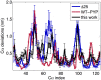
References
-
- Meyer TE, Tollin G, Hazzard JH, Cusanovich MA (1989) Photoactive yellow protein from the purple phototrophic bacterium, Ectothiorhodospira halophila. Quantum yield of photobleaching and effects of temperature, alcohols, glycerol, and sucrose on kinetics of photobleaching and recovery. Biophys J 56: 559–564. - PMC - PubMed
-
- Khan JS, Imamoto Y, Yamazaki Y, Kataoka M, Tokunaga F, et al. (2005) A Biosensor in the Time Domain Based on the Diffusion Coefficient Measurement: Intermolecular Interaction of an Intermediate of Photoactive Yellow Protein. Anal Chem 77: 6625–6629. - PubMed
-
- Ui M, Tanaka Y, Kinbara K (2012) Amplification of Light-induced Molecular-Shape Change by Supramolecular Machines. J Photopolym Sci Technol 25: 655–658.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

