Identification of a proximal progenitor population from murine fetal lungs with clonogenic and multilineage differentiation potential
- PMID: 25358791
- PMCID: PMC4223706
- DOI: 10.1016/j.stemcr.2014.07.010
Identification of a proximal progenitor population from murine fetal lungs with clonogenic and multilineage differentiation potential
Abstract
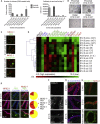
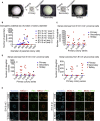
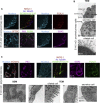
Lung development-associated diseases are major causes of morbidity and lethality in preterm infants and children. Access to the lung progenitor/stem cell populations controlling pulmonary development during embryogenesis and early postnatal years is essential to understand the molecular basis of such diseases. Using a Nkx2-1(mCherry) reporter mouse, we have identified and captured Nkx2-1-expressing lung progenitor cells from the proximal lung epithelium during fetal development. These cells formed clonal spheres in semisolid culture that could be maintained in vitro and demonstrated self-renewal and expansion capabilities over multiple passages. In-vitro-derived Nkx2-1-expressing clonal spheres differentiated into a polarized epithelium comprised of multiple cell lineages, including basal and secretory cells, that could repopulate decellularized lung scaffolds. Nkx2-1 expression thus defines a fetal lung epithelial progenitor cell population that can be used as a model system to study pulmonary development and associated pediatric diseases.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Broers J.L., de Leij L., Rot M.K., ter Haar A., Lane E.B., Leigh I.M., Wagenaar S.S., Vooijs G.P., Ramaekers F.C. Expression of intermediate filament proteins in fetal and adult human lung tissues. Differentiation. 1989;40:119–128. - PubMed
-
- Daniely Y., Liao G., Dixon D., Linnoila R.I., Lori A., Randell S.H., Oren M., Jetten A.M. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am. J. Physiol. Cell Physiol. 2004;287:C171–C181. - PubMed
-
- Delplanque A., Coraux C., Tirouvanziam R., Khazaal I., Puchelle E., Ambros P., Gaillard D., Péault B. Epithelial stem cell-mediated development of the human respiratory mucosa in SCID mice. J. Cell Sci. 2000;113:767–778. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

