Regulation of USP28 deubiquitinating activity by SUMO conjugation
- PMID: 25359778
- PMCID: PMC4263883
- DOI: 10.1074/jbc.M114.601849
Regulation of USP28 deubiquitinating activity by SUMO conjugation
Abstract
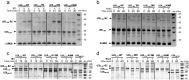
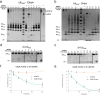
USP28 (ubiquitin-specific protease 28) is a deubiquitinating enzyme that has been implicated in the DNA damage response, the regulation of Myc signaling, and cancer progression. The half-life stability of major regulators of critical cellular pathways depends on the activities of specific ubiquitin E3 ligases that target them for proteosomal degradation and deubiquitinating enzymes that promote their stabilization. One function of the post-translational small ubiquitin modifier (SUMO) is the regulation of enzymatic activity of protein targets. In this work, we demonstrate that the SUMO modification of the N-terminal domain of USP28 negatively regulates its deubiquitinating activity, revealing a role for the N-terminal region as a regulatory module in the control of USP28 activity. Despite the presence of ubiquitin-binding domains in the N-terminal domain, its truncation does not impair deubiquitinating activity on diubiquitin or polyubiquitin chain substrates. In contrast to other characterized USP deubiquitinases, our results indicate that USP28 has a chain preference activity for Lys(11), Lys(48), and Lys(63) diubiquitin linkages.
Keywords: Proteolytic Enzyme; SUMO-interacting Motif (SIM); Sumoylation; Ubiquitin; Ubiquitin-dependent Protease.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 - PubMed
-
- Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 - PubMed
-
- Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

