Systematic transfer of prokaryotic sensors and circuits to mammalian cells
- PMID: 25360681
- PMCID: PMC4277766
- DOI: 10.1021/sb5002856
Systematic transfer of prokaryotic sensors and circuits to mammalian cells
Abstract
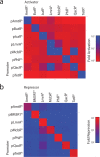
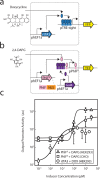
Prokaryotic regulatory proteins respond to diverse signals and represent a rich resource for building synthetic sensors and circuits. The TetR family contains >10(5) members that use a simple mechanism to respond to stimuli and bind distinct DNA operators. We present a platform that enables the transfer of these regulators to mammalian cells, which is demonstrated using human embryonic kidney (HEK293) and Chinese hamster ovary (CHO) cells. The repressors are modified to include nuclear localization signals (NLS) and responsive promoters are built by incorporating multiple operators. Activators are also constructed by modifying the protein to include a VP16 domain. Together, this approach yields 15 new regulators that demonstrate 19- to 551-fold induction and retain both the low levels of crosstalk in DNA binding specificity observed between the parent regulators in Escherichia coli, as well as their dynamic range of activity. By taking advantage of the DAPG small molecule sensing mediated by the PhlF repressor, we introduce a new inducible system with 50-fold induction and a threshold of 0.9 μM DAPG, which is comparable to the classic Dox-induced TetR system. A set of NOT gates is constructed from the new repressors and their response function quantified. Finally, the Dox- and DAPG- inducible systems and two new activators are used to build a synthetic enhancer (fuzzy AND gate), requiring the coordination of 5 transcription factors organized into two layers. This work introduces a generic approach for the development of mammalian genetic sensors and circuits to populate a toolbox that can be applied to diverse applications from biomanufacturing to living therapeutics.
Keywords: 2,4-diacetylphloroglucinol (DAPG); eukaryote; inducible system; mammalian synthetic biology; systems biology.
Figures


References
-
- Slusarczyk A. L.; Lin A.; Weiss R. (2012) Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 13, 406–420. - PubMed
-
- Fussenegger M.; Schlatter S.; Datwyler D.; Mazur X.; Bailey J. E. (1998) Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat. Biotechnol. 16, 468–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

