Direct interaction between Id1 and Zrf1 controls neural differentiation of embryonic stem cells
- PMID: 25361733
- PMCID: PMC4304729
- DOI: 10.15252/embr.201439560
Direct interaction between Id1 and Zrf1 controls neural differentiation of embryonic stem cells
Abstract
Id proteins are dominant-negative regulators within the HLH family of proteins. In embryonic stem cells (ESCs), Id1 and Id3 maintain the pluripotent state by preventing neural differentiation. The Id1-interacting protein Zrf1 plays a crucial role as a chromatin-bound factor in specification of the neural fate from ESCs. Here, we show that Id1 blocks Zrf1 recruitment to chromatin, thus preventing the activation of neural genes in ESCs. Upon differentiation, Id1 expression decreases thus inducing Zrf1 binding to neural genes. Importantly, depletion of Zrf1 rescues the expression of Polycomb targets involved in neural specification which are up-regulated in Id1 knock-out ESCs. We therefore identified Zrf1 as transcriptional regulator of neural fate downstream of Id1 in ESCs.
Keywords: Id1; Polycomb; epigenetics; neural development.
© 2014 The Authors.
Figures

ESCs were grown in either self-renewal conditions (serum plus LIF) or differentiation conditions (knock-out serum replacement, KSR, plus retinoic acid, RA). Extracts were subjected to immunoprecipitation with an antibody against either Zrf1 or Id1. Levels of co-immunoprecipitation of Id1 with Zrf1 (left), or of Zrf1 with Id1 (right), were assessed by Western blot.
Zrf1 ChIPs were performed in ESCs transfected with an empty vector (Ctrl) or in Id1-Flag overexpressing ESCs. Zrf1-depleted ESCs were used as a control. Values are reported as percentage of input. Standard deviation (SD) is representative of three independent experiments. *P < 0.05 and **P < 0.01 (paired t-test) represent comparison with control ESCs at the corresponding time point.
mRNA fold-change of Zrf1 targets under differentiation conditions (24 h RA). SD is representative of three independent experiments. *P < 0.05; **P < 0.01 (paired t-test).
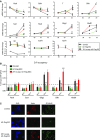
mRNA fold-changes of Id1, endogenous (mouse) and transgenic (human) Zrf1, pluripotent markers Cdh1 and Oct4, the neural markers Pax6, Sox1, Olig2, Hoxb1 and the epiblast/mesodermal marker T in control ESCs, Id1-Flag ESCs, or Id1-Flag ESCs transiently overexpressing Zrf1. Self-renewing conditions (day 0), and days 3 and 5 of neural differentiation were analyzed. SD is representative of three independent experiments. *P < 0.05 and **P < 0.01 (paired t-test) represent comparison with control ESCs at the corresponding time point.
ChIP analysis for Zrf1 in control ESCs, Id1-Flag ESCs, and Id1-Flag ESCs transiently overexpressing Zrf1, grown in self-renewing conditions (day 0) and during neural differentiation (day 5). SD is representative of three independent experiments. Dashed line represents IgG background. **P < 0.01 (paired t-test).
Immunostaining of the neural progenitor marker Nestin and of the post-mitotic neuronal marker β3 tubulin in control ESCs, Id1-Flag ESCs, and Id1-Flag ESCs transiently overexpressing Zrf1 at day 5 of neural differentiation.
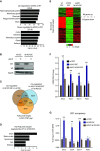
Gene ontology (GO) analysis of tissue expression of up- and down-regulated genes in Id1KO ESCs as compared to wild-type ESCs.
Western blot analysis of Id1 and Zrf1 expression in wild-type and Id1KO ESCs upon Zrf1 depletion.
Overlap between misregulated genes in Id1KO ESCs, genes significantly affected by Zrf1 depletion in Id1KO ESCs (identified by gene expression analysis), and Polycomb-bound genes in ESC (identified by ChIP-sequencing analysis).
GO analysis of tissue expression of 642 genes identified as Polycomb targets misregulated in Id1KO ESCs, which are significantly affected by Zrf1 depletion.
Heatmap analysis of Zrf1-regulated Polycomb genes whose expression is significantly enriched in brain and spinal cord.
mRNA fold-changes of Otx2, Pax6, Sox4, and Sox11 in control ESCs, Id1KO ESCs, and Zrf1-depleted Id1KO ESCs. SD is representative of three independent experiments. **P < 0.01 (paired t-test) represents comparison with Ctrl ESCs at the corresponding time point.
Zrf1 ChIPs at Otx2, Pax6, Sox4, and Sox11 promoters in control ESCs, Id1KO ESCs, and Zrf1-depleted Id1KO ESCs. SD is representative of three independent experiments. **P < 0.01 (paired t-test).
References
-
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. - PubMed
-
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. - PubMed
-
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. - PubMed
-
- Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. - PubMed
-
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–3905. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

