KnotProt: a database of proteins with knots and slipknots
- PMID: 25361973
- PMCID: PMC4383900
- DOI: 10.1093/nar/gku1059
KnotProt: a database of proteins with knots and slipknots
Abstract
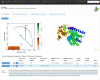
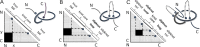
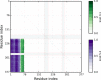
The protein topology database KnotProt, http://knotprot.cent.uw.edu.pl/, collects information about protein structures with open polypeptide chains forming knots or slipknots. The knotting complexity of the cataloged proteins is presented in the form of a matrix diagram that shows users the knot type of the entire polypeptide chain and of each of its subchains. The pattern visible in the matrix gives the knotting fingerprint of a given protein and permits users to determine, for example, the minimal length of the knotted regions (knot's core size) or the depth of a knot, i.e. how many amino acids can be removed from either end of the cataloged protein structure before converting it from a knot to a different type of knot. In addition, the database presents extensive information about the biological functions, families and fold types of proteins with non-trivial knotting. As an additional feature, the KnotProt database enables users to submit protein or polymer chains and generate their knotting fingerprints.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

