Mcph1/Brit1 deficiency promotes genomic instability and tumor formation in a mouse model
- PMID: 25362854
- PMCID: PMC4417661
- DOI: 10.1038/onc.2014.367
Mcph1/Brit1 deficiency promotes genomic instability and tumor formation in a mouse model
Abstract
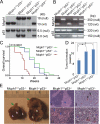
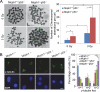
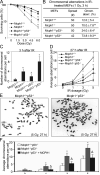
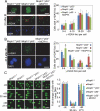
MCPH1, also known as BRIT1, has recently been identified as a novel key regulatory gene of the DNA damage response pathway. MCPH1 is located on human chromosome 8p23.1, where human cancers frequently show loss of heterozygosity. As such, MCPH1 is aberrantly expressed in many malignancies, including breast and ovarian cancers, and the function of MCPH1 has been implicated in tumor suppression. However, it remains poorly understood whether MCPH1 deficiency leads to tumorigenesis. Here we generated and studied both Mcph1(-/-) and Mcph1(-/-)p53(-/-) mice; we showed that Mcph1(-/-) mice developed tumors with long latency, and that primary lymphoma developed significantly earlier in Mcph1(-/-)p53(-/-) mice than in Mcph11(+/+)p53(-/-) and Mcph1(+/-)p53(-/-) mice. The Mcph1(-/-)p53(-/-) lymphomas and derived murine embryonic fibroblasts (MEFs) were both more sensitive to irradiation. Mcph1 deficiency resulted in remarkably increased chromosome and chromatid breaks in Mcph1(-/-)p53(-/-) lymphomas and MEFs, as determined by metaphase spread assay and spectral karyotyping analysis. In addition, Mcph1 deficiency significantly enhanced aneuploidy as well as abnormal centrosome multiplication in Mcph1(-/-)p53(-/-) cells. Meanwhile, Mcph1 deficiency impaired double strand break (DSB) repair in Mcph1(-/-)p53(-/-) MEFs as demonstrated by neutral Comet assay. Compared with Mcph1(+/+)p53(-/-) MEFs, homologous recombination and non-homologous end-joining activities were significantly decreased in Mcph1(-/-)p53(-/-) MEFs. Notably, reconstituted MCPH1 rescued the defects of DSB repair and alleviated chromosomal aberrations in Mcph1(-/-)p53(-/-) MEFs. Taken together, our data demonstrate MCPH1 deficiency promotes genomic instability and increases cancer susceptibility. Our study using knockout mouse models provides convincing genetic evidence that MCPH1 is a bona fide tumor suppressor gene. Its deficiency leading to defective DNA repair in tumors can be used to develop novel targeted cancer therapies in the future.
Figures

Similar articles
-
BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice.PLoS Genet. 2010 Jan 22;6(1):e1000826. doi: 10.1371/journal.pgen.1000826. PLoS Genet. 2010. PMID: 20107607 Free PMC article.
-
DNA damage response in microcephaly development of MCPH1 mouse model.DNA Repair (Amst). 2013 Aug;12(8):645-55. doi: 10.1016/j.dnarep.2013.04.017. Epub 2013 May 15. DNA Repair (Amst). 2013. PMID: 23683352
-
53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis.Proc Natl Acad Sci U S A. 2006 Feb 28;103(9):3310-5. doi: 10.1073/pnas.0511259103. Epub 2006 Feb 21. Proc Natl Acad Sci U S A. 2006. PMID: 16492765 Free PMC article.
-
Multiple roles of BRIT1/MCPH1 in DNA damage response, DNA repair, and cancer suppression.Yonsei Med J. 2010 May;51(3):295-301. doi: 10.3349/ymj.2010.51.3.295. Yonsei Med J. 2010. PMID: 20376879 Free PMC article. Review.
-
Aneuploidy and tumorigenesis.Semin Cell Dev Biol. 2011 Aug;22(6):595-601. doi: 10.1016/j.semcdb.2011.03.002. Epub 2011 Mar 15. Semin Cell Dev Biol. 2011. PMID: 21392584 Free PMC article. Review.
Cited by
-
Analysis of the "centrosome-ome" identifies MCPH1 deletion as a cause of centrosome amplification in human cancer.Sci Rep. 2020 Jul 17;10(1):11921. doi: 10.1038/s41598-020-68629-4. Sci Rep. 2020. PMID: 32681070 Free PMC article.
-
BRCT-domain protein BRIT1 influences class switch recombination.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8354-8359. doi: 10.1073/pnas.1708211114. Epub 2017 Jul 19. Proc Natl Acad Sci U S A. 2017. PMID: 28724724 Free PMC article.
-
Microcephalin 1/BRIT1-TRF2 interaction promotes telomere replication and repair, linking telomere dysfunction to primary microcephaly.Nat Commun. 2020 Nov 17;11(1):5861. doi: 10.1038/s41467-020-19674-0. Nat Commun. 2020. PMID: 33203878 Free PMC article.
-
BRIT1 dysfunction confers synergistic inhibition of hepatocellular carcinoma by targeting poly (ADP-ribose) polymerases and PI3K.Am J Cancer Res. 2020 Jun 1;10(6):1900-1918. eCollection 2020. Am J Cancer Res. 2020. PMID: 32642299 Free PMC article.
-
Silencing BRIT1 Facilitates the Abilities of Invasiveness and Migration in Trophoblast Cells.Med Sci Monit. 2018 Oct 19;24:7451-7458. doi: 10.12659/MSM.910229. Med Sci Monit. 2018. PMID: 30337515 Free PMC article.
References
-
- Venkatesh T, Suresh PS. Emerging roles of MCPH1: expedition from primary microcephaly to cancer. Eur J Cell Biol. 2014;93:98–105. - PubMed
-
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

