A positional Toll receptor code directs convergent extension in Drosophila
- PMID: 25363762
- PMCID: PMC4943584
- DOI: 10.1038/nature13953
A positional Toll receptor code directs convergent extension in Drosophila
Erratum in
-
Erratum: A positional Toll receptor code directs convergent extension in Drosophila.Nature. 2015 Nov 19;527(7578):398. doi: 10.1038/nature15719. Epub 2015 Oct 7. Nature. 2015. PMID: 26444242 No abstract available.
Abstract
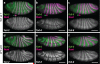
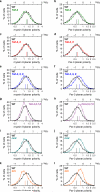
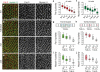
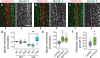
Elongation of the head-to-tail body axis by convergent extension is a conserved developmental process throughout metazoans. In Drosophila, patterns of transcription factor expression provide spatial cues that induce systematically oriented cell movements and promote tissue elongation. However, the mechanisms by which patterned transcriptional inputs control cell polarity and behaviour have long been elusive. We demonstrate that three Toll family receptors, Toll-2, Toll-6 and Toll-8, are expressed in overlapping transverse stripes along the anterior-posterior axis and act in combination to direct planar polarity and polarized cell rearrangements during convergent extension. Simultaneous disruption of all three receptors strongly reduces actomyosin-driven junctional remodelling and axis elongation, and an ectopic stripe of Toll receptor expression is sufficient to induce planar polarized actomyosin contractility. These results demonstrate that tissue-level patterns of Toll receptor expression provide spatial signals that link positional information from the anterior-posterior patterning system to the essential cell behaviours that drive convergent extension.
Figures







Comment in
-
Developmental biology: Polarize to elongate.Nature. 2014 Nov 27;515(7528):499-501. doi: 10.1038/nature13937. Epub 2014 Nov 5. Nature. 2014. PMID: 25363775 No abstract available.
References
-
- Zallen JA. Planar polarity and tissue morphogenesis. Cell. 2007;129:1051–1063. - PubMed
-
- Wallingford JB. Planar Cell Polarity and the Developmental Control of Cell Behavior in Vertebrate Embryos. Annu. Rev. Cell Dev. Biol. 2012;28:627–653. - PubMed
-
- Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 2012;28:687–717. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

