The mutational landscapes of genetic and chemical models of Kras-driven lung cancer
- PMID: 25363767
- PMCID: PMC4304785
- DOI: 10.1038/nature13898
The mutational landscapes of genetic and chemical models of Kras-driven lung cancer
Abstract
Next-generation sequencing of human tumours has refined our understanding of the mutational processes operative in cancer initiation and progression, yet major questions remain regarding the factors that induce driver mutations and the processes that shape mutation selection during tumorigenesis. Here we performed whole-exome sequencing on adenomas from three mouse models of non-small-cell lung cancer, which were induced either by exposure to carcinogens (methyl-nitrosourea (MNU) and urethane) or by genetic activation of Kras (Kras(LA2)). Although the MNU-induced tumours carried exactly the same initiating mutation in Kras as seen in the Kras(LA2) model (G12D), MNU tumours had an average of 192 non-synonymous, somatic single-nucleotide variants, compared with only six in tumours from the Kras(LA2) model. By contrast, the Kras(LA2) tumours exhibited a significantly higher level of aneuploidy and copy number alterations compared with the carcinogen-induced tumours, suggesting that carcinogen-induced and genetically engineered models lead to tumour development through different routes. The wild-type allele of Kras has been shown to act as a tumour suppressor in mouse models of non-small-cell lung cancer. We demonstrate that urethane-induced tumours from wild-type mice carry mostly (94%) Kras Q61R mutations, whereas those from Kras heterozygous animals carry mostly (92%) Kras Q61L mutations, indicating a major role for germline Kras status in mutation selection during initiation. The exome-wide mutation spectra in carcinogen-induced tumours overwhelmingly display signatures of the initiating carcinogen, while adenocarcinomas acquire additional C > T mutations at CpG sites. These data provide a basis for understanding results from human tumour genome sequencing, which has identified two broad categories of tumours based on the relative frequency of single-nucleotide variations and copy number alterations, and underline the importance of carcinogen models for understanding the complex mutation spectra seen in human cancers.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
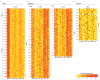
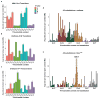



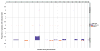

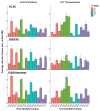



References
-
- Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- A12401/CRUK_/Cancer Research UK/United Kingdom
- U01 CA176287/CA/NCI NIH HHS/United States
- F31 CA180715/CA/NCI NIH HHS/United States
- 13031/CRUK_/Cancer Research UK/United Kingdom
- U01 CA176303/CA/NCI NIH HHS/United States
- A13031/CRUK_/Cancer Research UK/United Kingdom
- R01 CA184510/CA/NCI NIH HHS/United States
- U01 CA084244/CA/NCI NIH HHS/United States
- T32 GM007175/GM/NIGMS NIH HHS/United States
- 082356/WT_/Wellcome Trust/United Kingdom
- U01 CA84244/CA/NCI NIH HHS/United States
- U01 CA141455/CA/NCI NIH HHS/United States
- R01 CA111834/CA/NCI NIH HHS/United States
- T32GM007175/GM/NIGMS NIH HHS/United States
- UO1 CA176287/CA/NCI NIH HHS/United States
- F31 CA180669/CA/NCI NIH HHS/United States
- A14356/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

