Promoterless gene targeting without nucleases ameliorates haemophilia B in mice
- PMID: 25363772
- PMCID: PMC4297598
- DOI: 10.1038/nature13864
Promoterless gene targeting without nucleases ameliorates haemophilia B in mice
Abstract
Site-specific gene addition can allow stable transgene expression for gene therapy. When possible, this is preferred over the use of promiscuously integrating vectors, which are sometimes associated with clonal expansion and oncogenesis. Site-specific endonucleases that can induce high rates of targeted genome editing are finding increasing applications in biological discovery and gene therapy. However, two safety concerns persist: endonuclease-associated adverse effects, both on-target and off-target; and oncogene activation caused by promoter integration, even without nucleases. Here we perform recombinant adeno-associated virus (rAAV)-mediated promoterless gene targeting without nucleases and demonstrate amelioration of the bleeding diathesis in haemophilia B mice. In particular, we target a promoterless human coagulation factor IX (F9) gene to the liver-expressed mouse albumin (Alb) locus. F9 is targeted, along with a preceding 2A-peptide coding sequence, to be integrated just upstream to the Alb stop codon. While F9 is fused to Alb at the DNA and RNA levels, two separate proteins are synthesized by way of ribosomal skipping. Thus, F9 expression is linked to robust hepatic albumin expression without disrupting it. We injected an AAV8-F9 vector into neonatal and adult mice and achieved on-target integration into ∼0.5% of the albumin alleles in hepatocytes. We established that F9 was produced only from on-target integration, and ribosomal skipping was highly efficient. Stable F9 plasma levels at 7-20% of normal were obtained, and treated F9-deficient mice had normal coagulation times. In conclusion, transgene integration as a 2A-fusion to a highly expressed endogenous gene may obviate the requirement for nucleases and/or vector-borne promoters. This method may allow for safe and efficacious gene targeting in both infants and adults by greatly diminishing off-target effects while still providing therapeutic levels of expression from integration.
Conflict of interest statement
The authors declare no competing financial interests.
Conflict Statement: A.B. and M.A.K are founders of LogicBio Therapeutics, a startup biotechnology company with interests in the technology described in the manuscript.
Figures
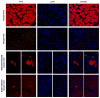

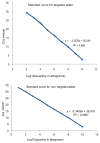
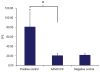
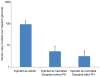
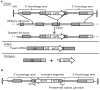

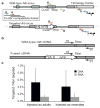

Comment in
-
Hitting the target without pulling the trigger.Mol Ther. 2015 Jan;23(1):4-6. doi: 10.1038/mt.2014.230. Mol Ther. 2015. PMID: 25561034 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

