Control of plant stem cell function by conserved interacting transcriptional regulators
- PMID: 25363783
- PMCID: PMC4297503
- DOI: 10.1038/nature13853
Control of plant stem cell function by conserved interacting transcriptional regulators
Abstract
Plant stem cells in the shoot apical meristem (SAM) and root apical meristem are necessary for postembryonic development of aboveground tissues and roots, respectively, while secondary vascular stem cells sustain vascular development. WUSCHEL (WUS), a homeodomain transcription factor expressed in the rib meristem of the Arabidopsis SAM, is a key regulatory factor controlling SAM stem cell populations, and is thought to establish the shoot stem cell niche through a feedback circuit involving the CLAVATA3 (CLV3) peptide signalling pathway. WUSCHEL-RELATED HOMEOBOX 5 (WOX5), which is specifically expressed in the root quiescent centre, defines quiescent centre identity and functions interchangeably with WUS in the control of shoot and root stem cell niches. WOX4, expressed in Arabidopsis procambial cells, defines the vascular stem cell niche. WUS/WOX family proteins are evolutionarily and functionally conserved throughout the plant kingdom and emerge as key actors in the specification and maintenance of stem cells within all meristems. However, the nature of the genetic regime in stem cell niches that centre on WOX gene function has been elusive, and molecular links underlying conserved WUS/WOX function in stem cell niches remain unknown. Here we demonstrate that the Arabidopsis HAIRY MERISTEM (HAM) family of transcription regulators act as conserved interacting cofactors with WUS/WOX proteins. HAM and WUS share common targets in vivo and their physical interaction is important in driving downstream transcriptional programs and in promoting shoot stem cell proliferation. Differences in the overlapping expression patterns of WOX and HAM family members underlie the formation of diverse stem cell niche locations, and the HAM family is essential for all of these stem cell niches. These findings establish a new framework for the control of stem cell production during plant development.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




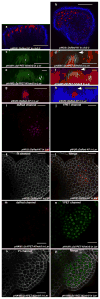

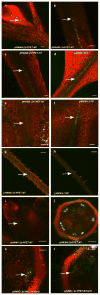
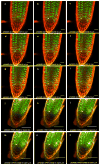
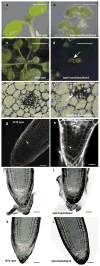




Comment in
-
Plant Development: Adding HAM to Stem Cell Control.Curr Biol. 2018 Nov 5;28(21):R1261-R1263. doi: 10.1016/j.cub.2018.09.039. Curr Biol. 2018. PMID: 30399352
References
-
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. - PubMed
-
- Sablowski R. The dynamic plant stem cell niches. Current opinion in plant biology. 2007;10:639–644. - PubMed
-
- Dinneny JR, Benfey PN. Plant stem cell niches: standing the test of time. Cell. 2008;132:553–557. - PubMed
-
- Laux T, Mayer KF, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

