High-throughput sequencing of human immunoglobulin variable regions with subtype identification
- PMID: 25364977
- PMCID: PMC4218849
- DOI: 10.1371/journal.pone.0111726
High-throughput sequencing of human immunoglobulin variable regions with subtype identification
Abstract
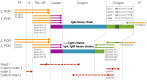
The humoral immune response plays a critical role in controlling infection, and the rapid adaptation to a broad range of pathogens depends on a highly diverse antibody repertoire. The advent of high-throughput sequencing technologies in the past decade has enabled insights into this immense diversity. However, not only the variable, but also the constant region of antibodies determines their in vivo activity. Antibody isotypes differ in effector functions and are thought to play a defining role in elicitation of immune responses, both in natural infection and in vaccination. We have developed an Illumina MiSeq high-throughput sequencing protocol that allows determination of the human IgG subtype alongside sequencing full-length antibody variable heavy chain regions. We thereby took advantage of the Illumina procedure containing two additional short reads as identifiers. By performing paired-end sequencing of the variable regions and customizing one of the identifier sequences to distinguish IgG subtypes, IgG transcripts with linked information of variable regions and IgG subtype can be retrieved. We applied our new method to the analysis of the IgG variable region repertoire from PBMC of an HIV-1 infected individual confirmed to have serum antibody reactivity to the Membrane Proximal External Region (MPER) of gp41. We found that IgG3 subtype frequencies in the memory B cell compartment increased after halted treatment and coincided with increased plasma antibody reactivity against the MPER domain. The sequencing strategy we developed is not restricted to analysis of IgG. It can be adopted for any Ig subtyping and beyond that for any research question where phasing of distant regions on the same amplicon is needed.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

