Regulation of the ahpC gene encoding alkyl hydroperoxide reductase in Mycobacterium smegmatis
- PMID: 25365321
- PMCID: PMC4218801
- DOI: 10.1371/journal.pone.0111680
Regulation of the ahpC gene encoding alkyl hydroperoxide reductase in Mycobacterium smegmatis
Abstract
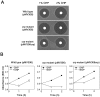
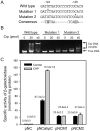
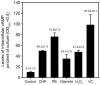
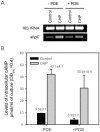
The ahpC (MSMEG_4891) gene encodes alkyl hydroperoxide reductase C in Mycobacterium smegmatis mc2155 and its expression is induced under oxidative stress conditions. Two well-defined inverted repeat sequences (IR1 and IR2) were identified in the upstream region of ahpC. Using a crp (cAMP receptor protein: MSMEG_6189) mutant and in vitro DNA-binding assay, it was demonstrated that the IR1 sequence serves as a Crp-binding site and that Crp functions as an activator in the regulation of ahpC expression. The expression level of ahpC was shown to be proportional to intracellular cAMP levels. Intracellular levels of cAMP were increased in M. smegmatis, when it was treated with oxidative stress inducers. The IR2 sequence is very similar to the known consensus sequence of FurA-binding sites and involved in the negative regulation of ahpC expression. Taken together, these results suggest that the induction of ahpC expression under oxidative stress conditions probably results from a combinatory effect of both inactivation of FurA by oxidative stress and activation of Crp in response to increased levels of cAMP.
Conflict of interest statement
Figures




References
-
- Wood ZA, Schroder E, Robin Harris J, Poole LB (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40. - PubMed
-
- Hillas PJ, del Alba FS, Oyarzabal J, Wilks A, Ortiz De Montellano PR (2000) The AhpC and AhpD antioxidant defense system of Mycobacterium tuberculosis . J Biol Chem 275: 18801–18809. - PubMed
-
- Bryk R, Griffin P, Nathan C (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407: 211–215. - PubMed
-
- Chen L, Xie QW, Nathan C (1998) Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol Cell 1: 795–805. - PubMed
-
- Master SS, Springer B, Sander P, Boettger EC, Deretic V, et al. (2002) Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148: 3139–3144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

