Histone methyltransferase Dot1L plays a role in postembryonic development in Xenopus tropicalis
- PMID: 25366346
- PMCID: PMC4314229
- DOI: 10.1096/fj.14-252171
Histone methyltransferase Dot1L plays a role in postembryonic development in Xenopus tropicalis
Abstract
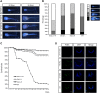
Histone methylations have been implicated to play important roles in diverse cellular processes. Of particular interest is the methylation of histone H3K79, which is catalyzed by an evolutionarily conserved methyltransferase, disruptor of telomeric silencing (Dot1)-like (Dot1L). To investigate the role of Dot1L during vertebrate development, we have generated a Dot1L-specific transcription activator-like effector nuclease (TALEN) nuclease to knockdown endogenous Dot1L in Xenopus tropicalis, a diploid species highly related to the well-known developmental model Xenopus laevis, a pseudotetraploid amphibian. We show that the TALEN was extremely efficient in mutating Dot1L when expressed in fertilized eggs, creating essentially Dot1L knockout embryos with little H3K79 methylation. Importantly, we observed that Dot1L knockdown had no apparent effect on embryogenesis because normally feeding tadpoles were formed, consistent with the lack of maternal Dot1L expression. On the other hand, Dot1L knockdown severely retarded the growth of the tadpoles and led to tadpole lethality prior to metamorphosis. These findings suggest that Dot1L and H3K79 methylation play an important role for tadpole growth and development prior to metamorphosis into a frog. Our findings further reveal interesting similarities and differences between Xenopus and mouse development and suggest the existence of 2 separate phases of vertebrate development with distinct requirements for epigenetic modifications.
Keywords: activation mark; epigenetics; histone modification; organogenesis.
© FASEB.
Figures



References
-
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 - PubMed
-
- Li B., Carey M., Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 - PubMed
-
- Barth T. K., Imhof A. (2010) Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 35, 618–626 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

