DNA repair mechanisms and human cytomegalovirus (HCMV) infection
- PMID: 25366712
- PMCID: PMC4429022
- DOI: 10.1007/s12223-014-0359-6
DNA repair mechanisms and human cytomegalovirus (HCMV) infection
Abstract
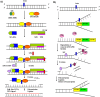
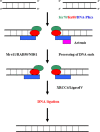
Herpesvirus infections, such as those induced by human cytomegalovirus (HCMV), induce specific DNA damages. DNA damages can lead to cell mutation, death, apoptosis and immune system activation. Various types of DNA damage are repaired through multiple repair pathways, such as base excision, nucleotide excision, homologous recombination and nonhomologous end joining. Changes in the activity of DNA repair proteins during viral infection can cause disturbances in the DNA repair system and change its mechanisms. This report reviews results from studies, assaying a DNA repair system in HCMV infection.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

