Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic cell MHCII expression for CD4 T cell priming during inflammation
- PMID: 25366967
- PMCID: PMC4235642
- DOI: 10.1084/jem.20140283
Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic cell MHCII expression for CD4 T cell priming during inflammation
Abstract
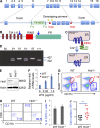
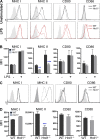
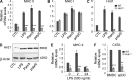
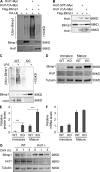
The ubiquitin pathway plays critical roles in antigen presentation. However, the ubiquitin ligases that regulate MHC gene transcription remain unidentified. We showed that the ubiquitin ligase Hrd1, expression of which is induced by Toll-like receptor (TLR) stimulation, is required for MHC-II but not MHC-I transcription in dendritic cells (DCs). Targeted Hrd1 gene deletion in DCs diminished MHC-II expression. As a consequence, Hrd1-null DCs failed to prime CD4(+) T cells without affecting the activation of CD8(+) T cells. Hrd1 catalyzed ubiquitination and degradation of the transcriptional suppressor B lymphocyte-induced maturation protein 1 (BLIMP1) to promote MHC-II expression. Genetic suppression of Hrd1 function in DCs protected mice from myelin oligodendrocyte glycoprotein (MOG)-induced experimental autoimmune encephalomyelitis (EAE). We identified Hrd1-mediated BLIMP1 ubiquitination as a previously unknown mechanism in programming DC for CD4(+) T cell activation during inflammation.
© 2014 Yang et al.
Figures



Similar articles
-
Loss of MHC II ubiquitination inhibits the activation and differentiation of CD4 T cells.Int Immunol. 2014 May;26(5):283-9. doi: 10.1093/intimm/dxt066. Epub 2013 Dec 26. Int Immunol. 2014. PMID: 24370470
-
Interferon-β regulates dendritic cell activation and migration in experimental autoimmune encephalomyelitis.Immunology. 2017 Nov;152(3):439-450. doi: 10.1111/imm.12781. Epub 2017 Jul 21. Immunology. 2017. PMID: 28646573 Free PMC article.
-
T cell-depleted splenocytes from mice pre-immunized with neuroantigen in incomplete Freund's adjuvant involved in protection from experimental autoimmune encephalomyelitis.Immunol Lett. 2014 Jan-Feb;157(1-2):38-44. doi: 10.1016/j.imlet.2013.11.001. Epub 2013 Nov 9. Immunol Lett. 2014. PMID: 24220208
-
Molecular mechanism and cellular function of MHCII ubiquitination.Immunol Rev. 2015 Jul;266(1):134-44. doi: 10.1111/imr.12303. Immunol Rev. 2015. PMID: 26085212 Free PMC article. Review.
-
Consequences of direct and indirect activation of dendritic cells on antigen presentation: functional implications and clinical considerations.Mol Immunol. 2013 Sep;55(2):175-8. doi: 10.1016/j.molimm.2012.10.034. Epub 2012 Nov 27. Mol Immunol. 2013. PMID: 23200227 Review.
Cited by
-
Autophagy Regulation by Crosstalk between miRNAs and Ubiquitination System.Int J Mol Sci. 2021 Nov 3;22(21):11912. doi: 10.3390/ijms222111912. Int J Mol Sci. 2021. PMID: 34769343 Free PMC article. Review.
-
Endoplasmic Reticulum Quality Control in Immune Cells.Front Cell Dev Biol. 2021 Sep 29;9:740653. doi: 10.3389/fcell.2021.740653. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34660599 Free PMC article. Review.
-
Increased cathepsin S in Prdm1-/- dendritic cells alters the TFH cell repertoire and contributes to lupus.Nat Immunol. 2017 Sep;18(9):1016-1024. doi: 10.1038/ni.3793. Epub 2017 Jul 10. Nat Immunol. 2017. PMID: 28692065 Free PMC article.
-
ER-associated degradation is required for vasopressin prohormone processing and systemic water homeostasis.J Clin Invest. 2017 Oct 2;127(10):3897-3912. doi: 10.1172/JCI94771. Epub 2017 Sep 18. J Clin Invest. 2017. PMID: 28920920 Free PMC article.
-
Ginsenoside Rb1 alleviates colitis in mice via activation of endoplasmic reticulum-resident E3 ubiquitin ligase Hrd1 signaling pathway.Acta Pharmacol Sin. 2021 Sep;42(9):1461-1471. doi: 10.1038/s41401-020-00561-9. Epub 2020 Dec 2. Acta Pharmacol Sin. 2021. PMID: 33268823 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

