Violation of the 12/23 rule of genomic V(D)J recombination is common in lymphocytes
- PMID: 25367293
- PMCID: PMC4315296
- DOI: 10.1101/gr.179770.114
Violation of the 12/23 rule of genomic V(D)J recombination is common in lymphocytes
Abstract
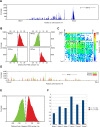
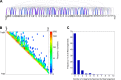
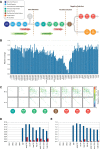
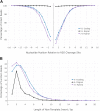
V(D)J genomic recombination joins single gene segments to encode an extensive repertoire of antigen receptor specificities in T and B lymphocytes. This process initiates with double-stranded breaks adjacent to conserved recombination signal sequences that contain either 12- or 23-nucleotide spacer regions. Only recombination between signal sequences with unequal spacers results in productive coding genes, a phenomenon known as the "12/23 rule." Here we present two novel genomic tools that allow the capture and analysis of immune locus rearrangements from whole thymic and splenic tissues using second-generation sequencing. Further, we provide strong evidence that the 12/23 rule of genomic recombination is frequently violated under physiological conditions, resulting in unanticipated hybrid recombinations in ∼10% of Tcra excision circles. Hence, we demonstrate that strict adherence to the 12/23 rule is intrinsic neither to recombination signal sequences nor to the catalytic process of recombination and propose that nonclassical excision circles are liberated during the formation of antigen receptor diversity.
© 2015 Parkinson et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Alexandre D, Chuchana P, Roncarolo MG, Yssel H, Spits H, Lefranc G, Lefranc MP. 1991. Reciprocal hybrid joints demonstrate successive V-J rearrangements on the same chromosome in the human TCRγ locus. Int Immunol 3: 973–982. - PubMed
-
- Bogue MA, Wang C, Zhu C, Roth DB. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7: 37–47. - PubMed
-
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. . 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442: 466–470. - PubMed
-
- Carroll AM, Slack JK, Mu X. 1993. V(D)J recombination generates a high frequency of nonstandard TCR Dδ-associated rearrangements in thymocytes. J Immunol 150: 2222–2230. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
