miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation
- PMID: 25367574
- PMCID: PMC4657560
- DOI: 10.1016/j.immuni.2014.09.015
miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation
Abstract
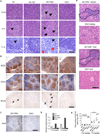
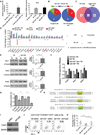
Chronic inflammation is a contributing factor to most life-shortening human diseases. However, the molecular and cellular mechanisms that sustain chronic inflammatory responses remain poorly understood, making it difficult to treat this deleterious condition. Using a mouse model of age-dependent inflammation that results from a deficiency in miR-146a, we demonstrate that miR-155 contributed to the progressive inflammatory disease that emerged as Mir146a(-/-) mice grew older. Upon analyzing lymphocytes from inflamed versus healthy middle-aged mice, we found elevated numbers of T follicular helper (Tfh) cells, germinal center (GC) B cells, and autoantibodies, all occurring in a miR-155-dependent manner. Further, Cd4-cre Mir155(fl/fl) mice were generated and demonstrated that miR-155 functions in T cells, in addition to its established role in B cells, to promote humoral immunity in a variety of contexts. Taken together, our study discovers that miR-146a and miR-155 counterregulate Tfh cell development that drives aberrant GC reactions during chronic inflammation.
Figures





Comment in
-
MicroRNAs in Tfh cells: micromanaging inflammaging.Immunity. 2014 Oct 16;41(4):509-11. doi: 10.1016/j.immuni.2014.10.003. Immunity. 2014. PMID: 25367565
References
-
- Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. The microRNA cluster miR-17 approximately 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840–848. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- K22 AI95375/AI/NIAID NIH HHS/United States
- DP2 GM111099/GM/NIGMS NIH HHS/United States
- R00 HL102228/HL/NHLBI NIH HHS/United States
- K08 CA133521/CA/NCI NIH HHS/United States
- DP2GM111099-01/DP/NCCDPHP CDC HHS/United States
- R56 AI107090/AI/NIAID NIH HHS/United States
- AI107090/AI/NIAID NIH HHS/United States
- R01CA166450-02/CA/NCI NIH HHS/United States
- R00HL102228-05/HL/NHLBI NIH HHS/United States
- R03NS070141/NS/NINDS NIH HHS/United States
- 5T32DK007115-39/DK/NIDDK NIH HHS/United States
- T32 DK007115/DK/NIDDK NIH HHS/United States
- R01 CA166450/CA/NCI NIH HHS/United States
- R03 NS070141/NS/NINDS NIH HHS/United States
- K22 AI095375/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

