Genomic and bioinformatic analysis of NADPH-cytochrome P450 reductase in Anopheles stephensi (Diptera: Culicidae)
- PMID: 25368081
- PMCID: PMC5443604
- DOI: 10.1093/jisesa/ieu027
Genomic and bioinformatic analysis of NADPH-cytochrome P450 reductase in Anopheles stephensi (Diptera: Culicidae)
Abstract
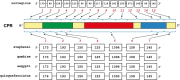
The cytochrome P450 monooxygenase (P450) enzyme system is a major mechanism of xenobiotic biotransformation. The nicotinamide adenine dinucleotide phosphate (NADPH)-cytochrome P450 reductase (CPR) is required for transfer of electrons from NADPH to P450. One CPR gene was identified in the genome of the malaria-transmitting mosquito Anopheles stephensi Liston (Diptera: Culicidae). The gene encodes a polypeptide containing highly conserved flavin mononucleotide-, flavin adenine dinucleotide-, and NADPH-binding domains, a unique characteristic of the reductase. Phylogenetic analysis revealed that the A. stephensi and other known mosquito CPRs belong to a monophyletic group distinctly separated from other insects in the same order, Diptera. Amino acid residues of CPRs involved in binding of P450 and cytochrome c are conserved between A. stephensi and the Norway rat Rattus norvegicus Berkenhout (Rodentia: Muridae). However, gene structure particularly within the coding region is evidently different between the two organisms. Such difference might arise during the evolution process as also seen in the difference of P450 families and isoforms found in these organisms. CPR in the mosquito A. stephensi is expected to be active and serve as an essential component of the P450 system.
Keywords: binding domain; gene structure; phylogenetic tree; sequence analysis.
© The Author 2014. Published by Oxford University Press on behalf of the Entomological Society of America.
Figures






Similar articles
-
Knockdown of NADPH-cytochrome P450 reductase results in reduced resistance to buprofezin in the small brown planthopper, Laodelphax striatellus (fallén).Pestic Biochem Physiol. 2016 Feb;127:21-7. doi: 10.1016/j.pestbp.2015.08.006. Epub 2015 Sep 5. Pestic Biochem Physiol. 2016. PMID: 26821654
-
Sequence characterization of cytochrome P450 CYP6P9 in pyrethroid resistant and susceptible Anopheles funestus (Diptera: Culicidae).Genet Mol Res. 2010 Mar 30;9(1):554-64. doi: 10.4238/vol9-1gmr719. Genet Mol Res. 2010. PMID: 20391340
-
Functional expression of mosquito NADPH-cytochrome P450 reductase in Escherichia coli.J Econ Entomol. 2007 Jun;100(3):946-53. doi: 10.1603/0022-0493(2007)100[946:feomnp]2.0.co;2. J Econ Entomol. 2007. PMID: 17598560
-
Identification and analysis of NADPH-cytochrome P450 reductase in Aedes sollicitans (Diptera: Culicidae).J Med Entomol. 2014 Sep;51(5):958-63. doi: 10.1603/me13072. J Med Entomol. 2014. PMID: 25276923
-
Clinical, structural and functional implications of mutations and polymorphisms in human NADPH P450 oxidoreductase.Fundam Clin Pharmacol. 2007 Aug;21(4):399-410. doi: 10.1111/j.1472-8206.2007.00520.x. Fundam Clin Pharmacol. 2007. PMID: 17635179 Review.
Cited by
-
Haplotype Diversity of NADPH-Cytochrome P450 Reductase Gene of Ophiocordyceps sinensis and the Effect on Fungal Infection in Host Insects.Microorganisms. 2020 Jun 29;8(7):968. doi: 10.3390/microorganisms8070968. Microorganisms. 2020. PMID: 32610431 Free PMC article.
-
Functional Validation of Endogenous Redox Partner Cytochrome P450 Reductase Reveals the Key P450s CYP6P9a/-b as Broad Substrate Metabolizers Conferring Cross-Resistance to Different Insecticide Classes in Anopheles funestus.Int J Mol Sci. 2024 Jul 25;25(15):8092. doi: 10.3390/ijms25158092. Int J Mol Sci. 2024. PMID: 39125661 Free PMC article.
References
-
- Black S. D., Coon M. J. . 1982. . Structural features of liver microsomal NADPH-cytochrome P450 reductase: hydrophobic domain, hydrophilic domain, and connecting region . J. Biol. Chem. 257 : 5929 – 5938 . - PubMed
-
- Dixit J., Srivastava H., Sharma M., Das M. K., Singh O. P., Raghavendra K., Nanda N., Dash A. P., Saksena D. N., Das A. . 2010. . Phylogenetic inference of Indian malaria vectors from multilocus DNA sequences . Infect. Genet. Evol. 10 : 755 – 763 . - PubMed
-
- Djouaka R. F., Bakare A. A., Coulibaly O. N., Akogbeto M. C., Ranson H., Hemingway J., Strode C. . 2008. . Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria . BMC Genomics 9 : 538 . - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

