EGR2 is critical for peripheral naïve T-cell differentiation and the T-cell response to influenza
- PMID: 25368162
- PMCID: PMC4246296
- DOI: 10.1073/pnas.1417215111
EGR2 is critical for peripheral naïve T-cell differentiation and the T-cell response to influenza
Abstract
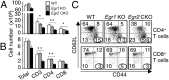
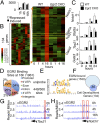
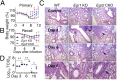
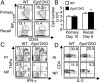
Early growth response 2 (EGR2) transcription factor negatively regulates T-cell activation, in contrast to the positive regulation of this process by EGR1. Here, we unexpectedly found that EGR2 promotes peripheral naïve T-cell differentiation, with delayed T-cell receptor-induced proliferation in naïve T cells from Egr2 conditional knockout (CKO) mice and decreased production of IFN-γ, IL-4, IL-9, and IL-17A in cells subjected to T-helper differentiation. Moreover, genes that promote T-cell activation, including Tbx21 and Notch1, had decreased expression in Egr2 CKO T cells and are direct EGR2 target genes. Following influenza infection, Egr2 CKO mice had delayed viral clearance, more weight loss, and more severe pathological changes in the lung than did WT and Egr1 KO mice, with decreased production of effector cytokines, increased infiltration of antigen-specific memory-precursor CD8(+) T cells, and lower numbers of lung-resident memory CD8(+) T cells. Thus, unexpectedly, EGR2 can function as a positive regulator that is essential for naïve T-cell differentiation and in vivo T-cell responses to a viral infection.
Keywords: EGR2; RNA-Seq; T cells; differentiation; influenza.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Local IL-4 expression in the lung reduces pulmonary influenza-virus-specific secondary cytotoxic T cell responses.Virology. 2000 Mar 30;269(1):66-77. doi: 10.1006/viro.2000.0187. Virology. 2000. PMID: 10725199
-
Temporal segregation of 4-1BB versus CD28-mediated costimulation: 4-1BB ligand influences T cell numbers late in the primary response and regulates the size of the T cell memory response following influenza infection.J Immunol. 2002 Apr 15;168(8):3777-85. doi: 10.4049/jimmunol.168.8.3777. J Immunol. 2002. PMID: 11937529
-
Egr2 and Egr3 in regulatory T cells cooperatively control systemic autoimmunity through Ltbp3-mediated TGF-β3 production.Proc Natl Acad Sci U S A. 2016 Dec 13;113(50):E8131-E8140. doi: 10.1073/pnas.1611286114. Epub 2016 Nov 30. Proc Natl Acad Sci U S A. 2016. PMID: 27911796 Free PMC article.
-
Double-negative T resident memory cells of the lung react to influenza virus infection via CD11c(hi) dendritic cells.Mucosal Immunol. 2016 Jul;9(4):999-1014. doi: 10.1038/mi.2015.91. Epub 2015 Sep 16. Mucosal Immunol. 2016. PMID: 26376363
-
Intranasal Delivery of Inactivated Influenza Virus and Poly(I:C) Adsorbed Corn-Based Nanoparticle Vaccine Elicited Robust Antigen-Specific Cell-Mediated Immune Responses in Maternal Antibody Positive Nursery Pigs.Front Immunol. 2020 Dec 16;11:596964. doi: 10.3389/fimmu.2020.596964. eCollection 2020. Front Immunol. 2020. PMID: 33391267 Free PMC article.
Cited by
-
Egr2 and 3 maintain anti-tumour responses of exhausted tumour infiltrating CD8 + T cells.Cancer Immunol Immunother. 2023 May;72(5):1139-1151. doi: 10.1007/s00262-022-03319-w. Epub 2022 Nov 7. Cancer Immunol Immunother. 2023. PMID: 36342511 Free PMC article.
-
Mechanisms underlying divergent responses of genetically distinct macrophages to IL-4.Sci Adv. 2021 Jun 16;7(25):eabf9808. doi: 10.1126/sciadv.abf9808. Print 2021 Jun. Sci Adv. 2021. PMID: 34134993 Free PMC article.
-
Early growth response 2, a novel target of pelvic organ prolapse, is highly expressed in anterior vaginal wall tissues with pelvic organ prolapse.Histochem Cell Biol. 2024 Feb;161(2):195-205. doi: 10.1007/s00418-023-02240-2. Epub 2023 Oct 24. Histochem Cell Biol. 2024. PMID: 37874337
-
Early Growth Response Gene 2-Expressing CD4+LAG3+ Regulatory T Cells: The Therapeutic Potential for Treating Autoimmune Diseases.Front Immunol. 2018 Feb 26;9:340. doi: 10.3389/fimmu.2018.00340. eCollection 2018. Front Immunol. 2018. PMID: 29535721 Free PMC article. Review.
-
Hidden in plain sight: The effects of BCG vaccination in the COVID-19 pandemic.J Med Virol. 2021 Apr;93(4):1950-1966. doi: 10.1002/jmv.26707. Epub 2020 Dec 17. J Med Virol. 2021. PMID: 33289122 Free PMC article. Review.
References
-
- Carleton M, et al. Early growth response transcription factors are required for development of CD4(-)CD8(-) thymocytes to the CD4(+)CD8(+) stage. J Immunol. 2002;168(4):1649–1658. - PubMed
-
- Bettini M, Xi H, Milbrandt J, Kersh GJ. Thymocyte development in early growth response gene 1-deficient mice. J Immunol. 2002;169(4):1713–1720. - PubMed
-
- Topilko P, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371(6500):796–799. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

