Protectome analysis: a new selective bioinformatics tool for bacterial vaccine candidate discovery
- PMID: 25368410
- PMCID: PMC4350036
- DOI: 10.1074/mcp.M114.039362
Protectome analysis: a new selective bioinformatics tool for bacterial vaccine candidate discovery
Abstract
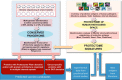
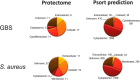
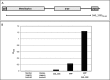
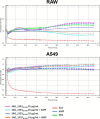
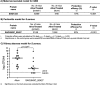
New generation vaccines are in demand to include only the key antigens sufficient to confer protective immunity among the plethora of pathogen molecules. In the last decade, large-scale genomics-based technologies have emerged. Among them, the Reverse Vaccinology approach was successfully applied to the development of an innovative vaccine against Neisseria meningitidis serogroup B, now available on the market with the commercial name BEXSERO® (Novartis Vaccines). The limiting step of such approaches is the number of antigens to be tested in in vivo models. Several laboratories have been trying to refine the original approach in order to get to the identification of the relevant antigens straight from the genome. Here we report a new bioinformatics tool that moves a first step in this direction. The tool has been developed by identifying structural/functional features recurring in known bacterial protective antigens, the so called "Protectome space," and using such "protective signatures" for protective antigen discovery. In particular, we applied this new approach to Staphylococcus aureus and Group B Streptococcus and we show that not only already known protective antigens were re-discovered, but also two new protective antigens were identified.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J.-F., Dougherty B. A., Merrick J. M., McKenney K., Sutton G., FitzHugh W., Fields C., Gocayne J. D., Scott J., Shirley R., Liu L.-l., Glodek A., Kelley J. M., Weidman J. F., Phillips C. A., Spriggs T., Hedblom E., Cotton M. D., Utterback T. R., Hanna M. C., Nguyen D. T., Saudek D. M., Brandon R. C., Fine L. D., Fritchman J. L., Fuhrmann J. L., Geoghagen N. S. M., Gnehm C. L., McDonald L. A., Small K. V., Fraser C. M., Smith H. O., Venter J. C., et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 - PubMed
-
- Pizza M., Scarlato V., Masignani V., Giuliani M. M., Aricó B., Comanducci M., Jennings G. T., Baldi L., Bartolini E., Capecchi B., Galeotti C. L., Luzzi E., Manetti R., Marchetti E., Mora M., Nuti S., Ratti G., Santini L., Savino S., Scarselli M., Storni E., Zuo P., Broeker M., Hundt E., Knapp B., Blair E., Mason T., Tettelin H., Hood D. W., Jeffries A. C., Saunders N. J., Granoff D. M., Venter J. C., Moxon E. R, Grandi G., Rappuoli R., et al. (2000) Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820 - PubMed
-
- Giuliani M. M., Adu-Bobie J., Comanducci M., Aricò B., Savino S., Santini L., Brunelli B., Bambini S., Biolchi A., Capecchi B., Cartocci E., Ciucchi L., Di Marcello F., Ferlicca F., Galli B., Luzzi E., Masignani V., Serruto D., Veggi D., Contorni M., Morandi M., Bartalesi A., Cinotti V., Mannucci D., Titta F., Ovidi E., Welsch J. A., Granoff D., Rappuoli R., Pizza M. (2006) A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U.S.A. 103, 10834–10839 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

