Multistep phosphorylation systems: tunable components of biological signaling circuits
- PMID: 25368420
- PMCID: PMC4230602
- DOI: 10.1091/mbc.E14-02-0774
Multistep phosphorylation systems: tunable components of biological signaling circuits
Abstract
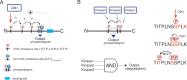
Multisite phosphorylation of proteins is a powerful signal processing mechanism that plays crucial roles in cell division and differentiation as well as in disease. We recently demonstrated a novel phenomenon in cell cycle regulation by showing that cyclin-dependent kinase-dependent multisite phosphorylation of a crucial substrate is performed sequentially in the N-to-C terminal direction along the disordered protein. The process is controlled by key parameters, including the distance between phosphorylation sites, the distribution of serines and threonines in sites, and the position of docking motifs. According to our model, linear patterns of phosphorylation along disordered protein segments determine the signal-response function of a multisite phosphorylation switch. Here we discuss the general advantages and engineering principles of multisite phosphorylation networks as processors of kinase signals. We also address the idea of using the mechanistic logic of linear multisite phosphorylation networks to design circuits for synthetic biology applications.
© 2014 Valk et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

References
-
- Ferrell JE., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–466. - PubMed
-
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

