Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells
- PMID: 25370495
- PMCID: PMC4222107
- DOI: 10.1128/mBio.02011-14
Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells
Abstract
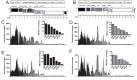
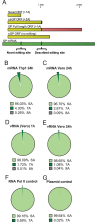
Deep sequencing of RNAs produced by Zaire ebolavirus (EBOV) or the Angola strain of Marburgvirus (MARV-Ang) identified novel viral and cellular mechanisms that diversify the coding and noncoding sequences of viral mRNAs and genomic RNAs. We identified previously undescribed sites within the EBOV and MARV-Ang mRNAs where apparent cotranscriptional editing has resulted in the addition of non-template-encoded residues within the EBOV glycoprotein (GP) mRNA, the MARV-Ang nucleoprotein (NP) mRNA, and the MARV-Ang polymerase (L) mRNA, such that novel viral translation products could be produced. Further, we found that the well-characterized EBOV GP mRNA editing site is modified at a high frequency during viral genome RNA replication. Additionally, editing hot spots representing sites of apparent adenosine deaminase activity were found in the MARV-Ang NP 3'-untranslated region. These studies identify novel filovirus-host interactions and reveal production of a greater diversity of filoviral gene products than was previously appreciated.
Importance: This study identifies novel mechanisms that alter the protein coding capacities of Ebola and Marburg virus mRNAs. Therefore, filovirus gene expression is more complex and diverse than previously recognized. These observations suggest new directions in understanding the regulation of filovirus gene expression.
Copyright © 2014 Shabman et al.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
