Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis
- PMID: 25370496
- PMCID: PMC4222108
- DOI: 10.1128/mBio.02020-14
Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis
Erratum in
- MBio. 2015;6(1):e02343-14
Abstract
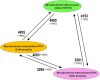
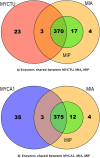
Mycobacterial evolution involves various processes, such as genome reduction, gene cooption, and critical gene acquisition. Our comparative genome size analysis of 44 mycobacterial genomes revealed that the nonpathogenic (NP) genomes were bigger than those of opportunistic (OP) or totally pathogenic (TP) mycobacteria, with the TP genomes being smaller yet variable in size--their genomic plasticity reflected their ability to evolve and survive under various environmental conditions. From the 44 mycobacterial species, 13 species, representing TP, OP, and NP, were selected for genomic-relatedness analyses. Analysis of homologous protein-coding genes shared between Mycobacterium indicus pranii (NP), Mycobacterium intracellulare ATCC 13950 (OP), and Mycobacterium tuberculosis H37Rv (TP) revealed that 4,995 (i.e., ~95%) M. indicaus pranii proteins have homology with M. intracellulare, whereas the homologies among M. indicus pranii, M. intracellulare ATCC 13950, and M. tuberculosis H37Rv were significantly lower. A total of 4,153 (~79%) M. indicus pranii proteins and 4,093 (~79%) M. intracellulare ATCC 13950 proteins exhibited homology with the M. tuberculosis H37Rv proteome, while 3,301 (~82%) and 3,295 (~82%) M. tuberculosis H37Rv proteins showed homology with M. indicus pranii and M. intracellulare ATCC 13950 proteomes, respectively. Comparative metabolic pathway analyses of TP/OP/NP mycobacteria showed enzymatic plasticity between M. indicus pranii (NP) and M. intracellulare ATCC 13950 (OP), Mycobacterium avium 104 (OP), and M. tuberculosis H37Rv (TP). Mycobacterium tuberculosis seems to have acquired novel alternate pathways with possible roles in metabolism, host-pathogen interactions, virulence, and intracellular survival, and by implication some of these could be potential drug targets.
Importance: The complete sequence analysis of Mycobacterium indicus pranii, a novel species of Mycobacterium shown earlier to have strong immunomodulatory properties and currently in use for the treatment of leprosy, places it evolutionarily at the point of transition to pathogenicity. With the purpose of establishing the importance of M. indicus pranii in providing insight into the virulence mechanism of tuberculous and nontuberculous mycobacteria, we carried out comparative genomic and proteomic analyses of 44 mycobacterial species representing nonpathogenic (NP), opportunistic (OP), and totally pathogenic (TP) mycobacteria. Our results clearly placed M. indicus pranii as an ancestor of the M. avium complex. Analyses of comparative metabolic pathways between M. indicus pranii (NP), M. tuberculosis (TP), and M. intracellulare (OP) pointed to the presence of novel alternative pathways in M. tuberculosis with implications for pathogenesis and survival in the human host and identification of new drug targets.
Copyright © 2014 Rahman et al.
Figures



Comment in
-
"Mycobacterium indicus pranii" is a strain of Mycobacterium intracellulare.mBio. 2015 Mar 3;6(2):e00013. doi: 10.1128/mBio.00013-15. mBio. 2015. PMID: 25736889 Free PMC article. No abstract available.
-
Reply to '"Mycobacterium indicus pranii" is a strain of Mycobacterium intracellulare': "M. indicus pranii" is a distinct strain, not derived from M. intracellulare, and is an organism at an evolutionary transition point between a fast grower and slow grower.mBio. 2015 Apr 7;6(2):e00352-15. doi: 10.1128/mBio.00352-15. mBio. 2015. PMID: 25852162 Free PMC article. No abstract available.
References
-
- Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99:3684–3689. 10.1073/pnas.052548299. - DOI - PMC - PubMed
-
- Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160. 10.1371/journal.ppat.1000160. - DOI - PMC - PubMed
-
- Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 6:95. 10.1186/1471-2148-6-95. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

