Structure-function analysis of heterodimer formation, oligomerization, and receptor binding of the Staphylococcus aureus bi-component toxin LukGH
- PMID: 25371205
- PMCID: PMC4281717
- DOI: 10.1074/jbc.M114.598110
Structure-function analysis of heterodimer formation, oligomerization, and receptor binding of the Staphylococcus aureus bi-component toxin LukGH
Abstract
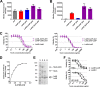
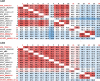
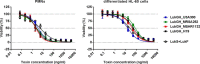
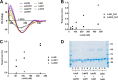
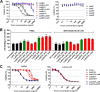
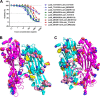
The bi-component leukocidins of Staphylococcus aureus are important virulence factors that lyse human phagocytic cells and contribute to immune evasion. The γ-hemolysins (HlgAB and HlgCB) and Panton-Valentine leukocidin (PVL or LukSF) were shown to assemble from soluble subunits into membrane-bound oligomers on the surface of target cells, creating barrel-like pore structures that lead to cell lysis. LukGH is the most distantly related member of this toxin family, sharing only 30-40% amino acid sequence identity with the others. We observed that, unlike other leukocidin subunits, recombinant LukH and LukG had low solubility and were unable to bind to target cells, unless both components were present. Using biolayer interferometry and intrinsic tryptophan fluorescence we detected binding of LukH to LukG in solution with an affinity in the low nanomolar range and dynamic light scattering measurements confirmed formation of a heterodimer. We elucidated the structure of LukGH by x-ray crystallography at 2.8-Å resolution. This revealed an octameric structure that strongly resembles that reported for HlgAB, but with important structural differences. Structure guided mutagenesis studies demonstrated that three salt bridges, not found in other bi-component leukocidins, are essential for dimer formation in solution and receptor binding. We detected weak binding of LukH, but not LukG, to the cellular receptor CD11b by biolayer interferometry, suggesting that in common with other members of this toxin family, the S-component has the primary contact role with the receptor. These new insights provide the basis for novel strategies to counteract this powerful toxin and Staphylococcus aureus pathogenesis.
Keywords: Bacterial Toxin; Crystal Structure; Crystallography; Microbial Pathogenesis; Mutagenesis; Staphylococcus aureus (S. aureus).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Spaan A. N., Surewaard B. G., Nijland R., van Strijp J. A. (2013) Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu. Rev. Microbiol. 67, 629–650 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

