A systematic survey of an intragenic epistatic landscape
- PMID: 25371431
- PMCID: PMC4271534
- DOI: 10.1093/molbev/msu301
A systematic survey of an intragenic epistatic landscape
Abstract
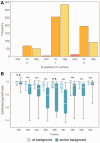
Mutations are the source of evolutionary variation. The interactions of multiple mutations can have important effects on fitness and evolutionary trajectories. We have recently described the distribution of fitness effects of all single mutations for a nine-amino-acid region of yeast Hsp90 (Hsp82) implicated in substrate binding. Here, we report and discuss the distribution of intragenic epistatic effects within this region in seven Hsp90 point mutant backgrounds of neutral to slightly deleterious effect, resulting in an analysis of more than 1,000 double mutants. We find negative epistasis between substitutions to be common, and positive epistasis to be rare--resulting in a pattern that indicates a drastic change in the distribution of fitness effects one step away from the wild type. This can be well explained by a concave relationship between phenotype and genotype (i.e., a concave shape of the local fitness landscape), suggesting mutational robustness intrinsic to the local sequence space. Structural analyses indicate that, in this region, epistatic effects are most pronounced when a solvent-inaccessible position is involved in the interaction. In contrast, all 18 observations of positive epistasis involved at least one mutation at a solvent-exposed position. By combining the analysis of evolutionary and biophysical properties of an epistatic landscape, these results contribute to a more detailed understanding of the complexity of protein evolution.
Keywords: distribution of fitness effects; epistasis; experimental evolution; fitness landscapes.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures





References
-
- Bateson W. Mendel’s principles of heredity. Cambridge (UK): Cambridge University Press; 1909. Available from: http://www.biodiversitylibrary.org/item/16926.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

