A novel member of glycoside hydrolase family 30 subfamily 8 with altered substrate specificity
- PMID: 25372685
- PMCID: PMC4722856
- DOI: 10.1107/S1399004714019531
A novel member of glycoside hydrolase family 30 subfamily 8 with altered substrate specificity
Abstract
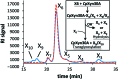
Endoxylanases classified into glycoside hydrolase family 30 subfamily 8 (GH30-8) are known to hydrolyze the hemicellulosic polysaccharide glucuronoxylan (GX) but not arabinoxylan or neutral xylooligosaccharides. This is owing to the specificity of these enzymes for the α-1,2-linked glucuronate (GA) appendage of GX. Limit hydrolysis of this substrate produces a series of aldouronates each containing a single GA substituted on the xylose penultimate to the reducing terminus. In this work, the structural and biochemical characterization of xylanase 30A from Clostridium papyrosolvens (CpXyn30A) is presented. This xylanase possesses a high degree of amino-acid identity to the canonical GH30-8 enzymes, but lacks the hallmark β8-α8 loop region which in part defines the function of this GH30 subfamily and its role in GA recognition. CpXyn30A is shown to have a similarly low activity on all xylan substrates, while hydrolysis of xylohexaose revealed a competing transglycosylation reaction. These findings are directly compared with the model GH30-8 enzyme from Bacillus subtilis, XynC. Despite its high sequence identity to the GH30-8 enzymes, CpXyn30A does not have any apparent specificity for the GA appendage. These findings confirm that the typically conserved β8-α8 loop region of these enzymes influences xylan substrate specificity but not necessarily β-1,4-xylanase function.
Keywords: glycoside hydrolase family 30; low-value biomass; wildfire; xylanase.
Figures




Similar articles
-
GH30 Glucuronoxylan-Specific Xylanase from Streptomyces turgidiscabies C56.Appl Environ Microbiol. 2018 Jan 31;84(4):e01850-17. doi: 10.1128/AEM.01850-17. Print 2018 Feb 15. Appl Environ Microbiol. 2018. PMID: 29180367 Free PMC article.
-
Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase.J Mol Biol. 2011 Mar 18;407(1):92-109. doi: 10.1016/j.jmb.2011.01.010. Epub 2011 Jan 19. J Mol Biol. 2011. PMID: 21256135
-
A plasmid borne, functionally novel glycoside hydrolase family 30 subfamily 8 endoxylanase from solventogenic Clostridium.Biochem J. 2018 May 4;475(9):1533-1551. doi: 10.1042/BCJ20180050. Biochem J. 2018. PMID: 29626157 Free PMC article.
-
Evidence for lysozyme-type mechanism of hydrolysis in xylanases.EXS. 1996;75:411-23. doi: 10.1007/978-3-0348-9225-4_20. EXS. 1996. PMID: 8765310 Review.
-
Xylanases of glycoside hydrolase family 30 - An overview.Biotechnol Adv. 2021 Mar-Apr;47:107704. doi: 10.1016/j.biotechadv.2021.107704. Epub 2021 Feb 3. Biotechnol Adv. 2021. PMID: 33548454 Review.
Cited by
-
A novel fungal GH30 xylanase with xylobiohydrolase auxiliary activity.Biotechnol Biofuels. 2019 May 11;12:120. doi: 10.1186/s13068-019-1455-2. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31110561 Free PMC article.
-
Structural Considerations on the Use of Endo-Xylanases for the Production of prebiotic Xylooligosaccharides from Biomass.Curr Protein Pept Sci. 2018;19(1):48-67. doi: 10.2174/1389203717666160923155209. Curr Protein Pept Sci. 2018. PMID: 27670134 Free PMC article. Review.
-
Crystal structure of GH30-7 endoxylanase C from the filamentous fungus Talaromyces cellulolyticus.Acta Crystallogr F Struct Biol Commun. 2020 Aug 1;76(Pt 8):341-349. doi: 10.1107/S2053230X20009024. Epub 2020 Jul 28. Acta Crystallogr F Struct Biol Commun. 2020. PMID: 32744245 Free PMC article.
-
The first crystal structure of a xylobiose-bound xylobiohydrolase with high functional specificity from the bacterial glycoside hydrolase family 30, subfamily 10.FEBS Lett. 2022 Sep;596(18):2449-2464. doi: 10.1002/1873-3468.14454. Epub 2022 Aug 4. FEBS Lett. 2022. PMID: 35876256 Free PMC article.
-
Endo-xylanases as tools for production of substituted xylooligosaccharides with prebiotic properties.Appl Microbiol Biotechnol. 2018 Nov;102(21):9081-9088. doi: 10.1007/s00253-018-9343-4. Epub 2018 Sep 8. Appl Microbiol Biotechnol. 2018. PMID: 30196329 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

