Enzyme-adenylate structure of a bacterial ATP-dependent DNA ligase with a minimized DNA-binding surface
- PMID: 25372693
- PMCID: PMC4220977
- DOI: 10.1107/S1399004714021099
Enzyme-adenylate structure of a bacterial ATP-dependent DNA ligase with a minimized DNA-binding surface
Abstract
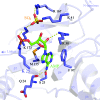
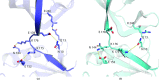
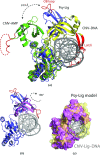
DNA ligases are a structurally diverse class of enzymes which share a common catalytic core and seal breaks in the phosphodiester backbone of double-stranded DNA via an adenylated intermediate. Here, the structure and activity of a recombinantly produced ATP-dependent DNA ligase from the bacterium Psychromonas sp. strain SP041 is described. This minimal-type ligase, like its close homologues, is able to ligate singly nicked double-stranded DNA with high efficiency and to join cohesive-ended and blunt-ended substrates to a more limited extent. The 1.65 Å resolution crystal structure of the enzyme-adenylate complex reveals no unstructured loops or segments, and suggests that this enzyme binds the DNA without requiring full encirclement of the DNA duplex. This is in contrast to previously characterized minimal DNA ligases from viruses, which use flexible loop regions for DNA interaction. The Psychromonas sp. enzyme is the first structure available for the minimal type of bacterial DNA ligases and is the smallest DNA ligase to be crystallized to date.
Keywords: ATP-dependent DNA ligase; Psychromonas sp. strain SP041.
Figures





Similar articles
-
DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases.Nucleic Acids Res. 2018 Sep 19;46(16):8616-8629. doi: 10.1093/nar/gky622. Nucleic Acids Res. 2018. PMID: 30007325 Free PMC article.
-
Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.J Biol Chem. 2019 Mar 29;294(13):5094-5104. doi: 10.1074/jbc.RA119.007445. Epub 2019 Feb 4. J Biol Chem. 2019. PMID: 30718283 Free PMC article.
-
Human DNA ligase I completely encircles and partially unwinds nicked DNA.Nature. 2004 Nov 25;432(7016):473-8. doi: 10.1038/nature03082. Nature. 2004. PMID: 15565146
-
DNA and RNA ligases: structural variations and shared mechanisms.Curr Opin Struct Biol. 2008 Feb;18(1):96-105. doi: 10.1016/j.sbi.2007.12.008. Epub 2008 Feb 8. Curr Opin Struct Biol. 2008. PMID: 18262407 Review.
-
Closing the gap on DNA ligase.Structure. 1996 Jun 15;4(6):653-6. doi: 10.1016/s0969-2126(96)00070-6. Structure. 1996. PMID: 8805556 Review.
Cited by
-
Using graphlet degree vectors to predict atomic displacement parameters in protein structures.Acta Crystallogr D Struct Biol. 2023 Dec 1;79(Pt 12):1109-1119. doi: 10.1107/S2059798323009142. Epub 2023 Nov 21. Acta Crystallogr D Struct Biol. 2023. PMID: 37987168 Free PMC article.
-
DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases.Nucleic Acids Res. 2018 Sep 19;46(16):8616-8629. doi: 10.1093/nar/gky622. Nucleic Acids Res. 2018. PMID: 30007325 Free PMC article.
-
Kinetics of T3-DNA Ligase-Catalyzed Phosphodiester Bond Formation Measured Using the α-Hemolysin Nanopore.ACS Nano. 2016 Dec 27;10(12):11127-11135. doi: 10.1021/acsnano.6b05995. Epub 2016 Dec 2. ACS Nano. 2016. PMID: 28024377 Free PMC article.
-
Temperature adaptation of DNA ligases from psychrophilic organisms.Extremophiles. 2019 May;23(3):305-317. doi: 10.1007/s00792-019-01082-y. Epub 2019 Mar 2. Extremophiles. 2019. PMID: 30826937
-
Structural insight into DNA joining: from conserved mechanisms to diverse scaffolds.Nucleic Acids Res. 2020 Sep 4;48(15):8225-8242. doi: 10.1093/nar/gkaa307. Nucleic Acids Res. 2020. PMID: 32365176 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

