Profiling status epilepticus-induced changes in hippocampal RNA expression using high-throughput RNA sequencing
- PMID: 25373493
- PMCID: PMC4894418
- DOI: 10.1038/srep06930
Profiling status epilepticus-induced changes in hippocampal RNA expression using high-throughput RNA sequencing
Abstract
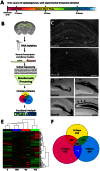
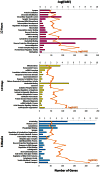
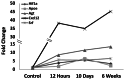
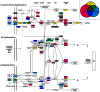
Status epilepticus (SE) is a life-threatening condition that can give rise to a number of neurological disorders, including learning deficits, depression, and epilepsy. Many of the effects of SE appear to be mediated by alterations in gene expression. To gain deeper insight into how SE affects the transcriptome, we employed the pilocarpine SE model in mice and Illumina-based high-throughput sequencing to characterize alterations in gene expression from the induction of SE, to the development of spontaneous seizure activity. While some genes were upregulated over the entire course of the pathological progression, each of the three sequenced time points (12-hour, 10-days and 6-weeks post-SE) had a largely unique transcriptional profile. Hence, genes that regulate synaptic physiology and transcription were most prominently altered at 12-hours post-SE; at 10-days post-SE, marked changes in metabolic and homeostatic gene expression were detected; at 6-weeks, substantial changes in the expression of cell excitability and morphogenesis genes were detected. At the level of cell signaling, KEGG analysis revealed dynamic changes within the MAPK pathways, as well as in CREB-associated gene expression. Notably, the inducible expression of several noncoding transcripts was also detected. These findings offer potential new insights into the cellular events that shape SE-evoked pathology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

