Molecular characterization of the monoclonal antibodies composing ZMAb: a protective cocktail against Ebola virus
- PMID: 25375093
- PMCID: PMC5381473
- DOI: 10.1038/srep06881
Molecular characterization of the monoclonal antibodies composing ZMAb: a protective cocktail against Ebola virus
Abstract
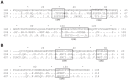
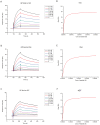
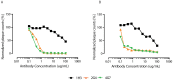
Ebola virus (EBOV) causes severe viral hemorrhagic fever in humans and non-human primates, with a case fatality rate of up to 88% in human outbreaks. Over the past 3 years, monoclonal antibody (mAb) cocktails have demonstrated high efficacy as treatments against EBOV infection. One such cocktail is ZMAb, which consists of three mouse antibodies, 1H3, 2G4, and 4G7. Here, we present the epitope binding properties of mAbs 1H3, 2G4, and 4G7. We showed that these antibodies have different variable region sequences, suggesting that the individual mAbs are not clonally related. All three antibodies were found to neutralize EBOV variant Mayinga. Additionally, 2G4 and 4G7 were shown to cross-inhibit each other in vitro and select for an escape mutation at the same position on the EBOV glycoprotein (GP), at amino acid 508. 1H3 selects an escape mutant at amino acid 273 on EBOV GP. Surface plasmon resonance studies showed that all three antibodies have dissociation constants on the order of 10(-7). In combination with previous studies evaluating the binding sites of other protective antibodies, our results suggest that antibodies targeting the GP1-GP2 interface and the glycan cap are often selected as efficacious antibodies for post-exposure interventions against EBOV.
Conflict of interest statement
Her Majesty the Queen in right of Canada holds a patent on the monoclonal antibodies 2G4, 4G7, and 1H3, PCT/CA2009/000070, “Monoclonal antibodies for Ebola and Marburg viruses.” The authors declare that they have no competing interests.
Figures


References
-
- Weissenhorn W., Carfí A., Lee K. H., Skehel J. J. & Wiley D. C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–16 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

