Co-dependence of HTLV-1 p12 and p8 functions in virus persistence
- PMID: 25375128
- PMCID: PMC4223054
- DOI: 10.1371/journal.ppat.1004454
Co-dependence of HTLV-1 p12 and p8 functions in virus persistence
Abstract
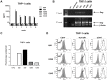
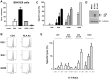
HTLV-1 orf-I is linked to immune evasion, viral replication and persistence. Examining the orf-I sequence of 160 HTLV-1-infected individuals; we found polymorphism of orf-I that alters the relative amounts of p12 and its cleavage product p8. Three groups were identified on the basis of p12 and p8 expression: predominantly p12, predominantly p8 and balanced expression of p12 and p8. We found a significant association between balanced expression of p12 and p8 with high viral DNA loads, a correlate of disease development. To determine the individual roles of p12 and p8 in viral persistence, we constructed infectious molecular clones expressing p12 and p8 (D26), predominantly p12 (G29S) or predominantly p8 (N26). As we previously showed, cells expressing N26 had a higher level of virus transmission in vitro. However, when inoculated into Rhesus macaques, cells producing N26 virus caused only a partial seroconversion in 3 of 4 animals and only 1 of those animals was HTLV-1 DNA positive by PCR. None of the animals exposed to G29S virus seroconverted or had detectable viral DNA. In contrast, 3 of 4 animals exposed to D26 virus seroconverted and were HTLV-1 positive by PCR. In vitro studies in THP-1 cells suggested that expression of p8 was sufficient for productive infection of monocytes. Since orf-I plays a role in T-cell activation and recognition; we compared the CTL response elicited by CD4+ T-cells infected with the different HTLV-1 clones. Although supernatant p19 levels and viral DNA loads for all four infected lines were similar, a significant difference in Tax-specific HLA.A2-restricted killing was observed. Cells infected with Orf-I-knockout virus (12KO), G29S or N26 were killed by CTLs, whereas cells infected with D26 virus were resistant to CTL killing. These results indicate that efficient viral persistence and spread require the combined functions of p12 and p8.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, et al. (1980) Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America 77: 7415–7419. - PMC - PubMed
-
- Gessain A, Barin F, Vernant J-C, Gout O, Maurs L, et al. (1985) Antibodies to human T-lymphotropic virus type I in patients with tropical spastic paraparesis. Lancet ii: 407–410. - PubMed
-
- Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, et al. (1986) HTLV-I associated myelopathy, a new clinical entity. Lancet 1: 1031–1032. - PubMed
-
- Franchini G, Lairmore MD (2007) Human T-cell leukemia/lymphoma virus types 1 and 2. In: Fields Virology. Philadelphia LWW, editor. pp. 2071–2106.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

