The CSN/COP9 signalosome regulates synaptonemal complex assembly during meiotic prophase I of Caenorhabditis elegans
- PMID: 25375142
- PMCID: PMC4222726
- DOI: 10.1371/journal.pgen.1004757
The CSN/COP9 signalosome regulates synaptonemal complex assembly during meiotic prophase I of Caenorhabditis elegans
Abstract
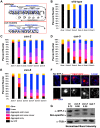
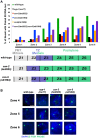
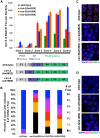
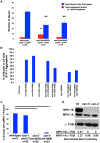
The synaptonemal complex (SC) is a conserved protein structure that holds homologous chromosome pairs together throughout much of meiotic prophase I. It is essential for the formation of crossovers, which are required for the proper segregation of chromosomes into gametes. The assembly of the SC is likely to be regulated by post-translational modifications. The CSN/COP9 signalosome has been shown to act in many pathways, mainly via the ubiquitin degradation/proteasome pathway. Here we examine the role of the CSN/COP9 signalosome in SC assembly in the model organism C. elegans. Our work shows that mutants in three subunits of the CSN/COP9 signalosome fail to properly assemble the SC. In these mutants, SC proteins aggregate, leading to a decrease in proper pairing between homologous chromosomes. The reduction in homolog pairing also results in an accumulation of recombination intermediates and defects in repair of meiotic DSBs to form the designated crossovers. The effect of the CSN/COP9 signalosome mutants on synapsis and crossover formation is due to increased neddylation, as reducing neddylation in these mutants can partially suppress their phenotypes. We also find a marked increase in apoptosis in csn mutants that specifically eliminates nuclei with aggregated SC proteins. csn mutants exhibit defects in germline proliferation, and an almost complete pachytene arrest due to an inability to activate the MAPK pathway. The work described here supports a previously unknown role for the CSN/COP9 signalosome in chromosome behavior during meiotic prophase I.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, et al. (2003) Synaptonemal Complex Assembly in C. elegans Is Dispensable for Loading Strand-Exchange Proteins but Critical for Proper Completion of Recombination. Developmental Cell 5: 463–474 10.1016/S1534-5807(03)00232-6 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

