Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila
- PMID: 25375361
- PMCID: PMC4222749
- DOI: 10.1371/journal.pgen.1004775
Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila
Abstract
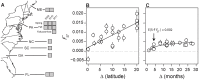
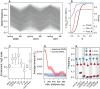
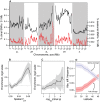
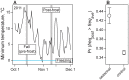
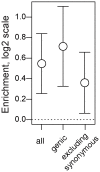
In many species, genomic data have revealed pervasive adaptive evolution indicated by the fixation of beneficial alleles. However, when selection pressures are highly variable along a species' range or through time adaptive alleles may persist at intermediate frequencies for long periods. So called "balanced polymorphisms" have long been understood to be an important component of standing genetic variation, yet direct evidence of the strength of balancing selection and the stability and prevalence of balanced polymorphisms has remained elusive. We hypothesized that environmental fluctuations among seasons in a North American orchard would impose temporally variable selection on Drosophila melanogaster that would drive repeatable adaptive oscillations at balanced polymorphisms. We identified hundreds of polymorphisms whose frequency oscillates among seasons and argue that these loci are subject to strong, temporally variable selection. We show that these polymorphisms respond to acute and persistent changes in climate and are associated in predictable ways with seasonally variable phenotypes. In addition, our results suggest that adaptively oscillating polymorphisms are likely millions of years old, with some possibly predating the divergence between D. melanogaster and D. simulans. Taken together, our results are consistent with a model of balancing selection wherein rapid temporal fluctuations in climate over generational time promotes adaptive genetic diversity at loci underlying polygenic variation in fitness related phenotypes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Gillespie JH (1973) Polymorphism in Random Environments. Theoretical Population Biology 4: 193–195.
-
- Ellner S, Sasaki A (1996) Patterns of genetic polymorphism maintained by fluctuating selection with overlapping generations. Theor Popul Biol 50: 31–65. - PubMed
-
- Korol AB, Kirzhner VM, Ronin YI, Nevo E (1996) Cyclical environmental changes as a factor maintaining genetic polymorphism. 2. Diploid selection for an additive trait. Evolution 50: 1432–1441. - PubMed
-
- Ewing EP (1979) Genetic-Variation in a Heterogeneous Environment. 7. Temporal and Spatial Heterogeneity in Infinite Populations. American Naturalist 114: 197–212.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

