RNA-seq analysis and de novo transcriptome assembly of Jerusalem artichoke (Helianthus tuberosus Linne)
- PMID: 25375764
- PMCID: PMC4222968
- DOI: 10.1371/journal.pone.0111982
RNA-seq analysis and de novo transcriptome assembly of Jerusalem artichoke (Helianthus tuberosus Linne)
Abstract
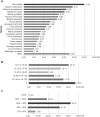
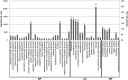
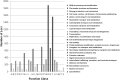
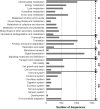
Jerusalem artichoke (Helianthus tuberosus L.) has long been cultivated as a vegetable and as a source of fructans (inulin) for pharmaceutical applications in diabetes and obesity prevention. However, transcriptomic and genomic data for Jerusalem artichoke remain scarce. In this study, Illumina RNA sequencing (RNA-Seq) was performed on samples from Jerusalem artichoke leaves, roots, stems and two different tuber tissues (early and late tuber development). Data were used for de novo assembly and characterization of the transcriptome. In total 206,215,632 paired-end reads were generated. These were assembled into 66,322 loci with 272,548 transcripts. Loci were annotated by querying against the NCBI non-redundant, Phytozome and UniProt databases, and 40,215 loci were homologous to existing database sequences. Gene Ontology terms were assigned to 19,848 loci, 15,434 loci were matched to 25 Clusters of Eukaryotic Orthologous Groups classifications, and 11,844 loci were classified into 142 Kyoto Encyclopedia of Genes and Genomes pathways. The assembled loci also contained 10,778 potential simple sequence repeats. The newly assembled transcriptome was used to identify loci with tissue-specific differential expression patterns. In total, 670 loci exhibited tissue-specific expression, and a subset of these were confirmed using RT-PCR and qRT-PCR. Gene expression related to inulin biosynthesis in tuber tissue was also investigated. Exsiting genetic and genomic data for H. tuberosus are scarce. The sequence resources developed in this study will enable the analysis of thousands of transcripts and will thus accelerate marker-assisted breeding studies and studies of inulin biosynthesis in Jerusalem artichoke.
Conflict of interest statement
Figures
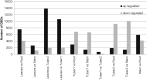

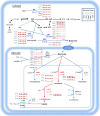
References
-
- Kosaric N, Cosentino GP, Wieczorek A, Duvnjak Z (1984) The Jerusalem artichoke as an agricultural crop. Biomass 5: 1–36.
-
- Monti A, Amaducci MT, Venturi G (2005) Growth response, leaf gas exchange and fructans accumulation of Jerusalem artichoke (Helianthus tuberosus L.) as affected by different water regimes. European Journal of Agronomy 23: 136–145.
-
- Yildiz GSP, Gungor T (2006) The effect of dietary Jerusalem artichoke (Helianthus tuberosus L.) on performance, egg quality characteristics and egg cholesterol content in laying hens. Czech J Anim Sci 51 (8): 349–354.
-
- Kays SJ, Nottingham SF (2008) Biology and Chemistry of Jerusalem Artichoke Helianthus tuberosus L. CRC press: 478.
-
- G DD B CB (1977) Irrigation, Fertilizer, Harvest Dates and Storage Effects on the Reducing Sugar and Fructose Concentrations of Jerusalem Artichoke Tubers. Can J Plant Sci. 591–596.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

