The evolution and genetics of virus host shifts
- PMID: 25375777
- PMCID: PMC4223060
- DOI: 10.1371/journal.ppat.1004395
The evolution and genetics of virus host shifts
Abstract
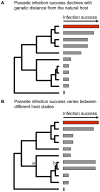
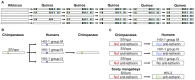
Emerging viral diseases are often the product of a host shift, where a pathogen jumps from its original host into a novel species. Phylogenetic studies show that host shifts are a frequent event in the evolution of most pathogens, but why pathogens successfully jump between some host species but not others is only just becoming clear. The susceptibility of potential new hosts can vary enormously, with close relatives of the natural host typically being the most susceptible. Often, pathogens must adapt to successfully infect a novel host, for example by evolving to use different cell surface receptors, to escape the immune response, or to ensure they are transmitted by the new host. In viruses there are often limited molecular solutions to achieve this, and the same sequence changes are often seen each time a virus infects a particular host. These changes may come at a cost to other aspects of the pathogen's fitness, and this may sometimes prevent host shifts from occurring. Here we examine how these evolutionary factors affect patterns of host shifts and disease emergence.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

