PINK1-mediated phosphorylation of Miro inhibits synaptic growth and protects dopaminergic neurons in Drosophila
- PMID: 25376463
- PMCID: PMC4223694
- DOI: 10.1038/srep06962
PINK1-mediated phosphorylation of Miro inhibits synaptic growth and protects dopaminergic neurons in Drosophila
Abstract
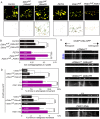
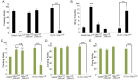
Mutations in the mitochondrial Ser/Thr kinase PINK1 cause Parkinson's disease. One of the substrates of PINK1 is the outer mitochondrial membrane protein Miro, which regulates mitochondrial transport. In this study, we uncovered novel physiological functions of PINK1-mediated phosphorylation of Miro, using Drosophila as a model. We replaced endogenous Drosophila Miro (DMiro) with transgenically expressed wildtype, or mutant DMiro predicted to resist PINK1-mediated phosphorylation. We found that the expression of phospho-resistant DMiro in a DMiro null mutant background phenocopied a subset of phenotypes of PINK1 null. Specifically, phospho-resistant DMiro increased mitochondrial movement and synaptic growth at larval neuromuscular junctions, and decreased the number of dopaminergic neurons in adult brains. Therefore, PINK1 may inhibit synaptic growth and protect dopaminergic neurons by phosphorylating DMiro. Furthermore, muscle degeneration, swollen mitochondria and locomotor defects found in PINK1 null flies were not observed in phospho-resistant DMiro flies. Thus, our study established an in vivo platform to define functional consequences of PINK1-mediated phosphorylation of its substrates.
Figures





References
-
- Valente E. M. et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160, 10.1126/science.1096284 (2004). - PubMed
-
- Vives-Bauza C., de Vries R. L., Tocilescu M. & Przedborski S. PINK1/Parkin direct mitochondria to autophagy. Autophagy 6, 315–316 (2010). - PubMed
-
- Whitworth A. J. & Pallanck L. J. The PINK1/Parkin pathway: a mitochondrial quality control system? J. Bioenerg. Biomembr. 41, 499–503, 10.1007/s10863-009-9253-3 (2009). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

