A Discovery Strategy for Selective Inhibitors of c-Src in Complex with the Focal Adhesion Kinase SH3/SH2-binding Region
- PMID: 25376742
- PMCID: PMC4444405
- DOI: 10.1111/cbdd.12473
A Discovery Strategy for Selective Inhibitors of c-Src in Complex with the Focal Adhesion Kinase SH3/SH2-binding Region
Abstract
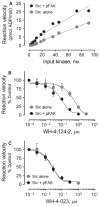
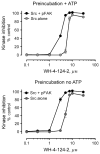
The c-Src tyrosine kinase co-operates with the focal adhesion kinase to regulate cell adhesion and motility. Focal adhesion kinase engages the regulatory SH3 and SH2 domains of c-Src, resulting in localized kinase activation that contributes to tumor cell metastasis. Using assay conditions where c-Src kinase activity required binding to a tyrosine phosphopeptide based on the focal adhesion kinase SH3-SH2 docking sequence, we screened a kinase-biased library for selective inhibitors of the Src/focal adhesion kinase peptide complex versus c-Src alone. This approach identified an aminopyrimidinyl carbamate compound, WH-4-124-2, with nanomolar inhibitory potency and fivefold selectivity for c-Src when bound to the phospho-focal adhesion kinase peptide. Molecular docking studies indicate that WH-4-124-2 may preferentially inhibit the 'DFG-out' conformation of the kinase active site. These findings suggest that interaction of c-Src with focal adhesion kinase induces a unique kinase domain conformation amenable to selective inhibition.
Keywords: SH2 domain; SH3 domain; Src kinase; cancer drug discovery; focal adhesion kinase; kinase inhibitors.
© 2014 John Wiley & Sons A/S.
Figures








References
-
- Brown MT, Cooper JA. Regulation, substrates, and functions of Src. Biochim Biophys Acta. 1996;1287:121–149. - PubMed
-
- Resh MD. Myristylation and palmitylation of Src family members: the fats of the matter. Cell. 1994;76:411–413. - PubMed
-
- Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. - PubMed
-
- Chong YP, Ia KK, Mulhern TD, Cheng HC. Endogenous and synthetic inhibitors of the Src-family protein tyrosine kinases. Biochim Biophys Acta. 2005;1754:210–220. - PubMed
-
- Chong YP, Chan AS, Chan KC, Williamson NA, Lerner EC, Smithgall TE, Bjorge JD, Fujita DJ, Purcell AW, Scholz G, Mulhern TD, Cheng HC. C-terminal Src kinase-homologous kinase (CHK), a unique inhibitor inactivating multiple active conformations of Src family tyrosine kinases. J Biol Chem. 2006;281:32988–32999. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

