The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis
- PMID: 25377552
- PMCID: PMC4302936
- DOI: 10.1242/dev.113639
The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis
Abstract
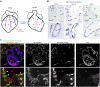
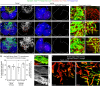
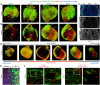
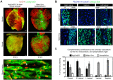
Identifying coronary artery progenitors and their developmental pathways could inspire novel regenerative treatments for heart disease. Multiple sources of coronary vessels have been proposed, including the sinus venosus (SV), endocardium and proepicardium, but their relative contributions to the coronary circulation and the molecular mechanisms regulating their development are poorly understood. We created an ApjCreER mouse line as a lineage-tracing tool to map SV-derived vessels onto the heart and compared the resulting lineage pattern with endocardial and proepicardial contributions to the coronary circulation. The data showed a striking compartmentalization to coronary development. ApjCreER-traced vessels contributed to a large number of arteries, capillaries and veins on the dorsal and lateral sides of the heart. By contrast, untraced vessels predominated in the midline of the ventral aspect and ventricular septum, which are vessel populations primarily derived from the endocardium. The proepicardium gave rise to a smaller fraction of vessels spaced relatively uniformly throughout the ventricular walls. Dorsal (SV-derived) and ventral (endocardial-derived) coronary vessels developed in response to different growth signals. The absence of VEGFC, which is expressed in the epicardium, dramatically inhibited dorsal and lateral coronary growth but left vessels on the ventral side unaffected. We propose that complementary SV-derived and endocardial-derived migratory routes unite to form the coronary vasculature and that the former requires VEGFC, revealing its role as a tissue-specific mediator of blood endothelial development.
Keywords: APLNR); Angiogenesis; Apelin receptor (APJ; Coronary vessel development; Endothelium; Sinus venosus; VEGF-C.
© 2014. Published by The Company of Biologists Ltd.
Figures



References
-
- Acharya A., Baek S. T., Huang G., Eskiocak B., Goetsch S., Sung C. Y., Banfi S., Sauer M. F., Olsen G. S., Duffield J. S. et al. (2012). The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 139, 2139-2149 10.1242/dev.079970 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

