Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins
- PMID: 25377555
- PMCID: PMC4302935
- DOI: 10.1242/dev.115048
Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins
Abstract
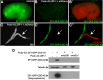
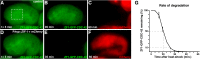
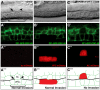
The capability to conditionally inactivate gene function is essential for understanding the molecular basis of development. In gene and mRNA targeting approaches, protein products can perdure, complicating genetic analysis. Current methods for selective protein degradation require drug treatment or take hours for protein removal, limiting their utility in studying rapid developmental processes in vivo. Here, we repurpose an endogenous protein degradation system to rapidly remove targeted C. elegans proteins. We show that upon expression of the E3 ubiquitin ligase substrate-recognition subunit ZIF-1, proteins tagged with the ZF1 zinc-finger domain can be quickly degraded in all somatic cell types examined with temporal and spatial control. We demonstrate that genes can be engineered to become conditional loss-of-function alleles by introducing sequences encoding the ZF1 tag into endogenous loci. Finally, we use ZF1 tagging to establish the site of cdc-42 gene function during a cell invasion event. ZF1 tagging provides a powerful new tool for the analysis of dynamic developmental events.
Keywords: C. elegans; Genetic tool; Mosaic; Protein degradation.
© 2014. Published by The Company of Biologists Ltd.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

