Severing of a hydrogen bond disrupts amino acid networks in the catalytically active state of the alpha subunit of tryptophan synthase
- PMID: 25377949
- PMCID: PMC4380980
- DOI: 10.1002/pro.2598
Severing of a hydrogen bond disrupts amino acid networks in the catalytically active state of the alpha subunit of tryptophan synthase
Abstract
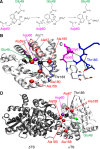
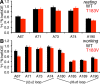
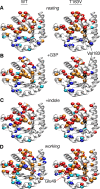
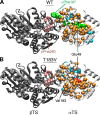
Conformational changes in the β2α2 and β6α6 loops in the alpha subunit of tryptophan synthase (αTS) are important for enzyme catalysis and coordinating substrate channeling with the beta subunit (βTS). It was previously shown that disrupting the hydrogen bond interactions between these loops through the T183V substitution on the β6α6 loop decreases catalytic efficiency and impairs substrate channeling. Results presented here also indicate that the T183V substitution decreases catalytic efficiency in Escherchia coli αTS in the absence of the βTS subunit. Nuclear magnetic resonance (NMR) experiments indicate that the T183V substitution leads to local changes in the structural dynamics of the β2α2 and β6α6 loops. We have also used NMR chemical shift covariance analyses (CHESCA) to map amino acid networks in the presence and absence of the T183V substitution. Under conditions of active catalytic turnover, the T183V substitution disrupts long-range networks connecting the catalytic residue Glu49 to the αTS-βTS binding interface, which might be important in the coordination of catalytic activities in the tryptophan synthase complex. The approach that we have developed here will likely find general utility in understanding long-range impacts on protein structure and dynamics of amino acid substitutions generated through protein engineering and directed evolution approaches, and provide insight into disease and drug-resistance mutations.
Keywords: amino acid networks; chemical shift covariance analysis; enzyme mechanisms; nuclear magnetic resonance; protein dynamics; tryptophan synthase.
© 2014 The Protein Society.
Figures



References
-
- Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J Mol Biol. 2004;344:1135–1146. - PubMed
-
- Bode C, Kovacs IA, Szalay MS, Palotai R, Korcsmaros T, Csermely P. Network analysis of protein dynamics. FEBS Lett. 2007;581:2776–2782. - PubMed
-
- Csermely P, Sandhu KS, Hazai E, Hoksza Z, Kiss HJ, Miozzo F, Veres DV, Piazza F, Nussinov R. Disordered proteins and network disorder in network descriptions of protein structure, dynamics and function: hypotheses and a comprehensive review. Curr Protein Pept Sci. 2012;13:19–33. - PubMed
-
- Lee J, Goodey NM. Catalytic contributions from remote regions of enzyme structure. Chem Rev. 2011;111:7595–7624. - PubMed
-
- Reynolds KA, Russ WP, Socolich M, Ranganathan R. Evolution-based design of proteins. Methods Enzymol. 2013;523:213–235. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

