Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes
- PMID: 25378320
- PMCID: PMC4245946
- DOI: 10.1093/nar/gku1077
Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes
Abstract
The cold shock domain is one of the most highly conserved motifs between bacteria and higher eukaryotes. Y-box-binding proteins represent a subfamily of cold shock domain proteins with pleiotropic functions, ranging from transcription in the nucleus to translation in the cytoplasm. These proteins have been investigated in all major model organisms except Caenorhabditis elegans. In this study, we set out to fill this gap and present a functional characterization of CEYs, the C. elegans Y-box-binding proteins. We find that, similar to other organisms, CEYs are essential for proper gametogenesis. However, we also report a novel function of these proteins in the formation of large polysomes in the soma. In the absence of the somatic CEYs, polysomes are dramatically reduced with a simultaneous increase in monosomes and disomes, which, unexpectedly, has no obvious impact on animal biology. Because transcripts that are enriched in polysomes in wild-type animals tend to be less abundant in the absence of CEYs, our findings suggest that large polysomes might depend on transcript stabilization mediated by CEY proteins.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

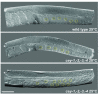
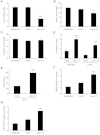
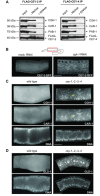
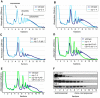
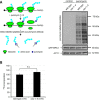
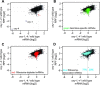

References
-
- Sommerville J. Activities of cold-shock domain proteins in translation control. Bioessays. 1999;21:319–325. - PubMed
-
- Eliseeva I.A., Kim E.R., Guryanov S.G., Ovchinnikov L.P., Lyabin D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Moscow) 2011;76:1402–1433. - PubMed
-
- Ruzanov P.V., Evdokimova V.M., Korneeva N.L., Hershey J.W., Ovchinnikov L.P. Interaction of the universal mRNA-binding protein, p50, with actin: a possible link between mRNA and microfilaments. J. Cell Sci. 1999;112:3487–3496. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

